|
Symbol Name ID |
Kat8
K(lysine) acetyltransferase 8 MGI:1915023 |
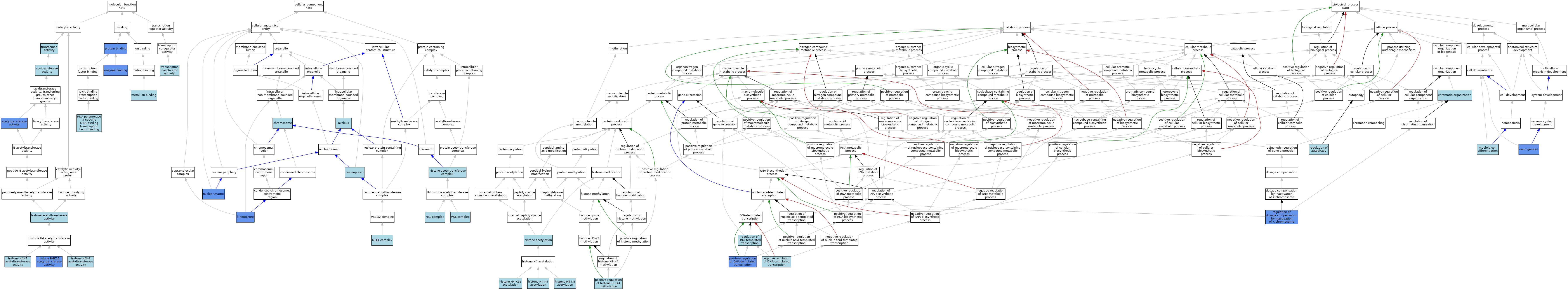
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0016407 | acetyltransferase activity | IDA | J:150958 | |||||||||
| Molecular Function | GO:0016746 | acyltransferase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0019899 | enzyme binding | IPI | J:150958 | |||||||||
| Molecular Function | GO:0004402 | histone acetyltransferase activity | IEA | J:72247 | |||||||||
| Molecular Function | GO:0004402 | histone acetyltransferase activity | IEA | J:72245 | |||||||||
| Molecular Function | GO:0046972 | histone H4K16 acetyltransferase activity | IMP | J:297798 | |||||||||
| Molecular Function | GO:0046972 | histone H4K16 acetyltransferase activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0046972 | histone H4K16 acetyltransferase activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0043995 | histone H4K5 acetyltransferase activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0043996 | histone H4K8 acetyltransferase activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0046872 | metal ion binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:120330 | |||||||||
| Molecular Function | GO:0061629 | RNA polymerase II-specific DNA-binding transcription factor binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0003713 | transcription coactivator activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0016740 | transferase activity | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005694 | chromosome | IEA | J:60000 | |||||||||
| Cellular Component | GO:0000123 | histone acetyltransferase complex | ISO | J:164563 | |||||||||
| Cellular Component | GO:0000776 | kinetochore | IDA | J:86777 | |||||||||
| Cellular Component | GO:0071339 | MLL1 complex | ISO | J:164563 | |||||||||
| Cellular Component | GO:0072487 | MSL complex | ISO | J:164563 | |||||||||
| Cellular Component | GO:0044545 | NSL complex | ISO | J:164563 | |||||||||
| Cellular Component | GO:0016363 | nuclear matrix | IDA | J:174530 | |||||||||
| Cellular Component | GO:0005654 | nucleoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005634 | nucleus | ISO | J:164563 | |||||||||
| Biological Process | GO:0006325 | chromatin organization | IEA | J:60000 | |||||||||
| Biological Process | GO:0016573 | histone acetylation | IEA | J:72247 | |||||||||
| Biological Process | GO:0043984 | histone H4-K16 acetylation | ISO | J:164563 | |||||||||
| Biological Process | GO:0043981 | histone H4-K5 acetylation | ISO | J:164563 | |||||||||
| Biological Process | GO:0043982 | histone H4-K8 acetylation | ISO | J:164563 | |||||||||
| Biological Process | GO:0030099 | myeloid cell differentiation | ISO | J:164563 | |||||||||
| Biological Process | GO:0045892 | negative regulation of DNA-templated transcription | ISO | J:164563 | |||||||||
| Biological Process | GO:0022008 | neurogenesis | IMP | J:297798 | |||||||||
| Biological Process | GO:0022008 | neurogenesis | ISO | J:164563 | |||||||||
| Biological Process | GO:0045893 | positive regulation of DNA-templated transcription | ISO | J:164563 | |||||||||
| Biological Process | GO:0045893 | positive regulation of DNA-templated transcription | IMP | J:313850 | |||||||||
| Biological Process | GO:0045893 | positive regulation of DNA-templated transcription | NAS | J:313850 | |||||||||
| Biological Process | GO:0051571 | positive regulation of histone H3-K4 methylation | ISO | J:164563 | |||||||||
| Biological Process | GO:0010506 | regulation of autophagy | ISO | J:73065 | |||||||||
| Biological Process | GO:0006355 | regulation of DNA-templated transcription | IEA | J:72247 | |||||||||
| Biological Process | GO:1900095 | regulation of dosage compensation by inactivation of X chromosome | IDA | J:313850 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/16/2024 MGI 6.23 |

|
|
|
||