|
Symbol Name ID |
Fbxo32
F-box protein 32 MGI:1914981 |
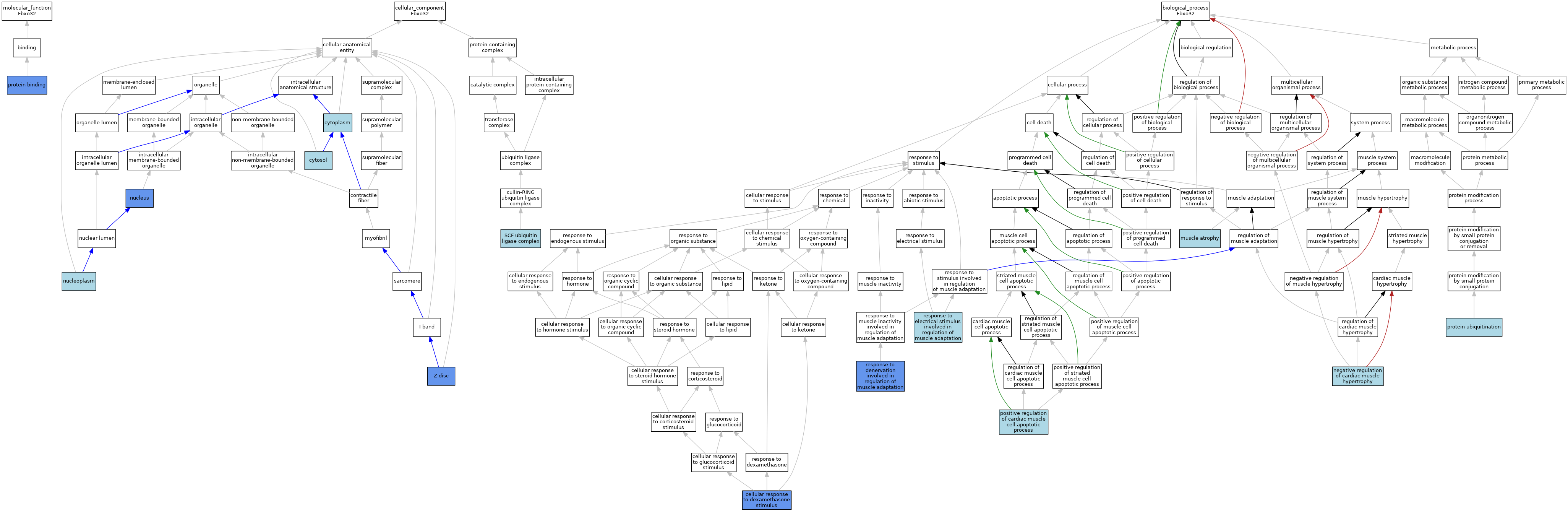
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0005515 | protein binding | IPI | J:172547 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005829 | cytosol | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005829 | cytosol | TAS | Reactome:R-NUL-9624625 | |||||||||
| Cellular Component | GO:0005654 | nucleoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IDA | J:177547 | |||||||||
| Cellular Component | GO:0005634 | nucleus | ISS | J:73126 | |||||||||
| Cellular Component | GO:0019005 | SCF ubiquitin ligase complex | ISO | J:164563 | |||||||||
| Cellular Component | GO:0019005 | SCF ubiquitin ligase complex | IBA | J:265628 | |||||||||
| Cellular Component | GO:0030018 | Z disc | IDA | J:172547 | |||||||||
| Biological Process | GO:0071549 | cellular response to dexamethasone stimulus | IDA | J:199171 | |||||||||
| Biological Process | GO:0014889 | muscle atrophy | ISO | J:155856 | |||||||||
| Biological Process | GO:0010614 | negative regulation of cardiac muscle hypertrophy | ISO | J:155856 | |||||||||
| Biological Process | GO:0010666 | positive regulation of cardiac muscle cell apoptotic process | ISO | J:155856 | |||||||||
| Biological Process | GO:0016567 | protein ubiquitination | ISO | J:164563 | |||||||||
| Biological Process | GO:0016567 | protein ubiquitination | IBA | J:265628 | |||||||||
| Biological Process | GO:0014894 | response to denervation involved in regulation of muscle adaptation | IDA | J:200498 | |||||||||
| Biological Process | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation | ISO | J:155856 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/16/2024 MGI 6.23 |

|
|
|
||