|
Symbol Name ID |
Rpl11
ribosomal protein L11 MGI:1914275 |
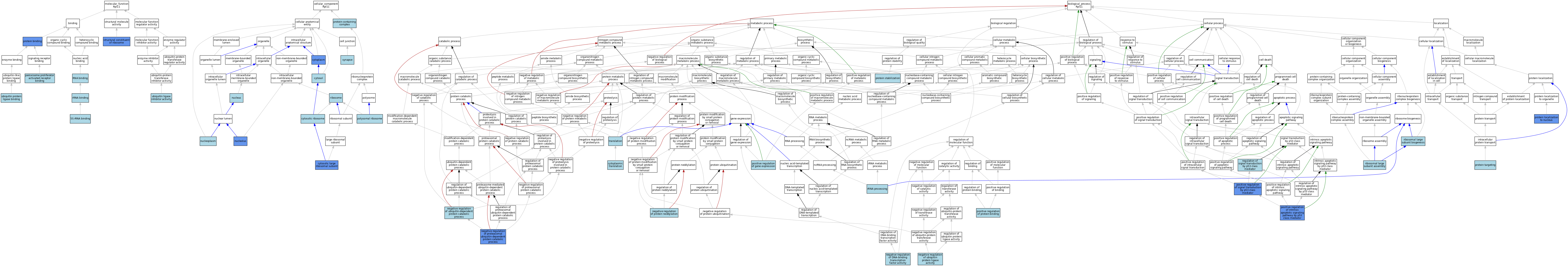
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0008097 | 5S rRNA binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0042975 | peroxisome proliferator activated receptor binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:240382 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:241062 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:187617 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:164200 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:169789 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:126004 | |||||||||
| Molecular Function | GO:0003723 | RNA binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:0019843 | rRNA binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0003735 | structural constituent of ribosome | ISO | J:164563 | |||||||||
| Molecular Function | GO:0003735 | structural constituent of ribosome | IDA | J:333015 | |||||||||
| Molecular Function | GO:0003735 | structural constituent of ribosome | IBA | J:265628 | |||||||||
| Molecular Function | GO:1990948 | ubiquitin ligase inhibitor activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0031625 | ubiquitin protein ligase binding | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | NAS | J:314083 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IDA | J:241062 | |||||||||
| Cellular Component | GO:0005829 | cytosol | TAS | Reactome:R-MMU-9634671 | |||||||||
| Cellular Component | GO:0022625 | cytosolic large ribosomal subunit | IPI | J:314083 | |||||||||
| Cellular Component | GO:0022625 | cytosolic large ribosomal subunit | IDA | J:333015 | |||||||||
| Cellular Component | GO:0022625 | cytosolic large ribosomal subunit | ISO | J:155856 | |||||||||
| Cellular Component | GO:0022625 | cytosolic large ribosomal subunit | ISO | J:164563 | |||||||||
| Cellular Component | GO:0022625 | cytosolic large ribosomal subunit | IBA | J:265628 | |||||||||
| Cellular Component | GO:0022626 | cytosolic ribosome | ISO | J:155856 | |||||||||
| Cellular Component | GO:0022626 | cytosolic ribosome | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005730 | nucleolus | IDA | J:169789 | |||||||||
| Cellular Component | GO:0005730 | nucleolus | IDA | J:241062 | |||||||||
| Cellular Component | GO:0005730 | nucleolus | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005654 | nucleoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IEA | J:60000 | |||||||||
| Cellular Component | GO:0042788 | polysomal ribosome | ISO | J:164563 | |||||||||
| Cellular Component | GO:0032991 | protein-containing complex | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005840 | ribosome | IEA | J:72247 | |||||||||
| Cellular Component | GO:0005840 | ribosome | IEA | J:60000 | |||||||||
| Cellular Component | GO:0045202 | synapse | ISO | J:155856 | |||||||||
| Biological Process | GO:0002181 | cytoplasmic translation | ISO | J:164563 | |||||||||
| Biological Process | GO:0002181 | cytoplasmic translation | NAS | J:314083 | |||||||||
| Biological Process | GO:0043433 | negative regulation of DNA-binding transcription factor activity | ISO | J:155856 | |||||||||
| Biological Process | GO:0032435 | negative regulation of proteasomal ubiquitin-dependent protein catabolic process | IMP | J:241062 | |||||||||
| Biological Process | GO:2000435 | negative regulation of protein neddylation | ISO | J:164563 | |||||||||
| Biological Process | GO:1904667 | negative regulation of ubiquitin protein ligase activity | ISO | J:164563 | |||||||||
| Biological Process | GO:2000059 | negative regulation of ubiquitin-dependent protein catabolic process | ISO | J:164563 | |||||||||
| Biological Process | GO:0010628 | positive regulation of gene expression | ISO | J:164563 | |||||||||
| Biological Process | GO:1902255 | positive regulation of intrinsic apoptotic signaling pathway by p53 class mediator | IMP | J:240382 | |||||||||
| Biological Process | GO:0032092 | positive regulation of protein binding | ISO | J:164563 | |||||||||
| Biological Process | GO:1901798 | positive regulation of signal transduction by p53 class mediator | IMP | J:241062 | |||||||||
| Biological Process | GO:0034504 | protein localization to nucleus | IMP | J:169789 | |||||||||
| Biological Process | GO:0050821 | protein stabilization | ISO | J:164563 | |||||||||
| Biological Process | GO:0006605 | protein targeting | ISO | J:164563 | |||||||||
| Biological Process | GO:1901796 | regulation of signal transduction by p53 class mediator | ISO | J:164563 | |||||||||
| Biological Process | GO:0000027 | ribosomal large subunit assembly | ISO | J:164563 | |||||||||
| Biological Process | GO:0042273 | ribosomal large subunit biogenesis | ISO | J:164563 | |||||||||
| Biological Process | GO:0006364 | rRNA processing | ISO | J:164563 | |||||||||
| Biological Process | GO:0006412 | translation | IEA | J:72247 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 05/07/2024 MGI 6.23 |

|
|
|
||