|
Symbol Name ID |
Psmd14
proteasome (prosome, macropain) 26S subunit, non-ATPase, 14 MGI:1913284 |
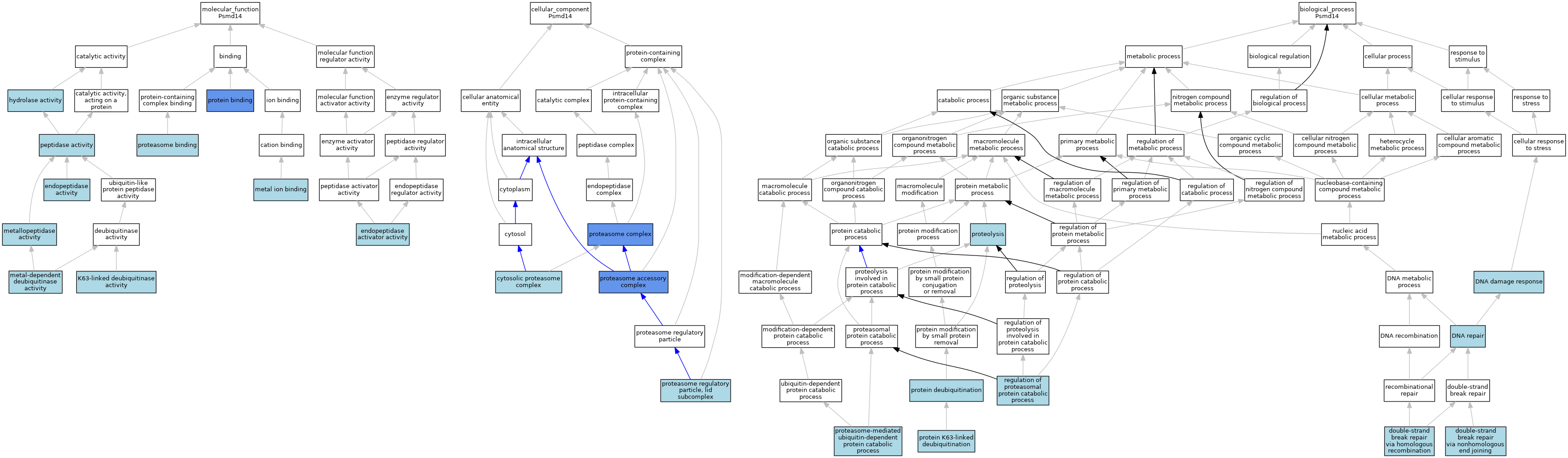
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0061133 | endopeptidase activator activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0004175 | endopeptidase activity | ISS | J:64935 | |||||||||
| Molecular Function | GO:0016787 | hydrolase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0061578 | K63-linked deubiquitinase activity | IEA | J:72247 | |||||||||
| Molecular Function | GO:0046872 | metal ion binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0140492 | metal-dependent deubiquitinase activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0008237 | metallopeptidase activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0008233 | peptidase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0070628 | proteasome binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0070628 | proteasome binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:200421 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:112707 | |||||||||
| Cellular Component | GO:0031597 | cytosolic proteasome complex | ISO | J:155856 | |||||||||
| Cellular Component | GO:0022624 | proteasome accessory complex | IDA | J:188902 | |||||||||
| Cellular Component | GO:0000502 | proteasome complex | ISO | J:164563 | |||||||||
| Cellular Component | GO:0000502 | proteasome complex | ISS | J:64935 | |||||||||
| Cellular Component | GO:0000502 | proteasome complex | IDA | J:112707 | |||||||||
| Cellular Component | GO:0008541 | proteasome regulatory particle, lid subcomplex | ISO | J:164563 | |||||||||
| Cellular Component | GO:0008541 | proteasome regulatory particle, lid subcomplex | IBA | J:265628 | |||||||||
| Biological Process | GO:0006974 | DNA damage response | IEA | J:60000 | |||||||||
| Biological Process | GO:0006281 | DNA repair | IEA | J:60000 | |||||||||
| Biological Process | GO:0000724 | double-strand break repair via homologous recombination | ISO | J:164563 | |||||||||
| Biological Process | GO:0006303 | double-strand break repair via nonhomologous end joining | ISO | J:164563 | |||||||||
| Biological Process | GO:0043161 | proteasome-mediated ubiquitin-dependent protein catabolic process | IBA | J:265628 | |||||||||
| Biological Process | GO:0016579 | protein deubiquitination | IBA | J:265628 | |||||||||
| Biological Process | GO:0070536 | protein K63-linked deubiquitination | ISO | J:164563 | |||||||||
| Biological Process | GO:0006508 | proteolysis | IEA | J:60000 | |||||||||
| Biological Process | GO:0061136 | regulation of proteasomal protein catabolic process | ISO | J:164563 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/30/2024 MGI 6.23 |

|
|
|
||