|
Symbol Name ID |
Akirin2
akirin 2 MGI:1889364 |
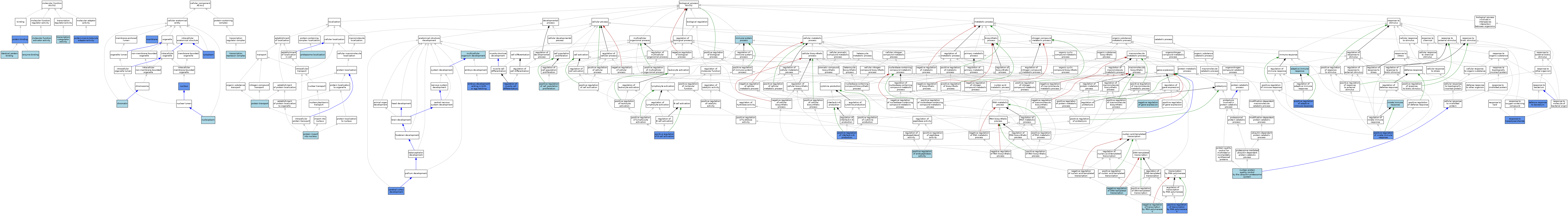
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0019899 | enzyme binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0042802 | identical protein binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0140677 | molecular function activator activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:216064 | |||||||||
| Molecular Function | GO:0030674 | protein-macromolecule adaptor activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0030674 | protein-macromolecule adaptor activity | IDA | J:216064 | |||||||||
| Molecular Function | GO:0003712 | transcription coregulator activity | IBA | J:265628 | |||||||||
| Cellular Component | GO:0000785 | chromatin | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IDA | J:275243 | |||||||||
| Cellular Component | GO:0016020 | membrane | IDA | J:275243 | |||||||||
| Cellular Component | GO:0005654 | nucleoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005634 | nucleus | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IDA | J:275243 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IDA | J:216064 | |||||||||
| Cellular Component | GO:0005634 | nucleus | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005634 | nucleus | ISO | J:130487 | |||||||||
| Cellular Component | GO:0017053 | transcription repressor complex | ISO | J:155856 | |||||||||
| Biological Process | GO:0002250 | adaptive immune response | IEA | J:60000 | |||||||||
| Biological Process | GO:0021987 | cerebral cortex development | IMP | J:237195 | |||||||||
| Biological Process | GO:0042742 | defense response to bacterium | IMP | J:216064 | |||||||||
| Biological Process | GO:0009792 | embryo development ending in birth or egg hatching | IMP | J:130487 | |||||||||
| Biological Process | GO:0002376 | immune system process | IEA | J:60000 | |||||||||
| Biological Process | GO:0045087 | innate immune response | IEA | J:60000 | |||||||||
| Biological Process | GO:0007275 | multicellular organism development | IEA | J:60000 | |||||||||
| Biological Process | GO:0045892 | negative regulation of DNA-templated transcription | ISO | J:155856 | |||||||||
| Biological Process | GO:0010629 | negative regulation of gene expression | ISO | J:155856 | |||||||||
| Biological Process | GO:0000122 | negative regulation of transcription by RNA polymerase II | ISO | J:155856 | |||||||||
| Biological Process | GO:0071630 | nuclear protein quality control by the ubiquitin-proteasome system | ISO | J:164563 | |||||||||
| Biological Process | GO:0002821 | positive regulation of adaptive immune response | IMP | J:322367 | |||||||||
| Biological Process | GO:0050871 | positive regulation of B cell activation | IMP | J:322367 | |||||||||
| Biological Process | GO:0008284 | positive regulation of cell population proliferation | ISO | J:155856 | |||||||||
| Biological Process | GO:0010950 | positive regulation of endopeptidase activity | ISO | J:155856 | |||||||||
| Biological Process | GO:0045089 | positive regulation of innate immune response | IMP | J:216064 | |||||||||
| Biological Process | GO:0045089 | positive regulation of innate immune response | IBA | J:265628 | |||||||||
| Biological Process | GO:0032755 | positive regulation of interleukin-6 production | IDA | J:216064 | |||||||||
| Biological Process | GO:0032755 | positive regulation of interleukin-6 production | IMP | J:130487 | |||||||||
| Biological Process | GO:0045944 | positive regulation of transcription by RNA polymerase II | IDA | J:216064 | |||||||||
| Biological Process | GO:0045944 | positive regulation of transcription by RNA polymerase II | IBA | J:265628 | |||||||||
| Biological Process | GO:0045944 | positive regulation of transcription by RNA polymerase II | IMP | J:130487 | |||||||||
| Biological Process | GO:0045944 | positive regulation of transcription by RNA polymerase II | IMP | J:130487 | |||||||||
| Biological Process | GO:0045944 | positive regulation of transcription by RNA polymerase II | IMP | J:130487 | |||||||||
| Biological Process | GO:0045944 | positive regulation of transcription by RNA polymerase II | IMP | J:130487 | |||||||||
| Biological Process | GO:0031144 | proteasome localization | ISO | J:164563 | |||||||||
| Biological Process | GO:0006606 | protein import into nucleus | ISO | J:164563 | |||||||||
| Biological Process | GO:0015031 | protein transport | IEA | J:60000 | |||||||||
| Biological Process | GO:0051147 | regulation of muscle cell differentiation | IMP | J:275243 | |||||||||
| Biological Process | GO:0032496 | response to lipopolysaccharide | IMP | J:130487 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/16/2024 MGI 6.23 |

|
|
|
||