|
Symbol Name ID |
Slc12a5
solute carrier family 12, member 5 MGI:1862037 |
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
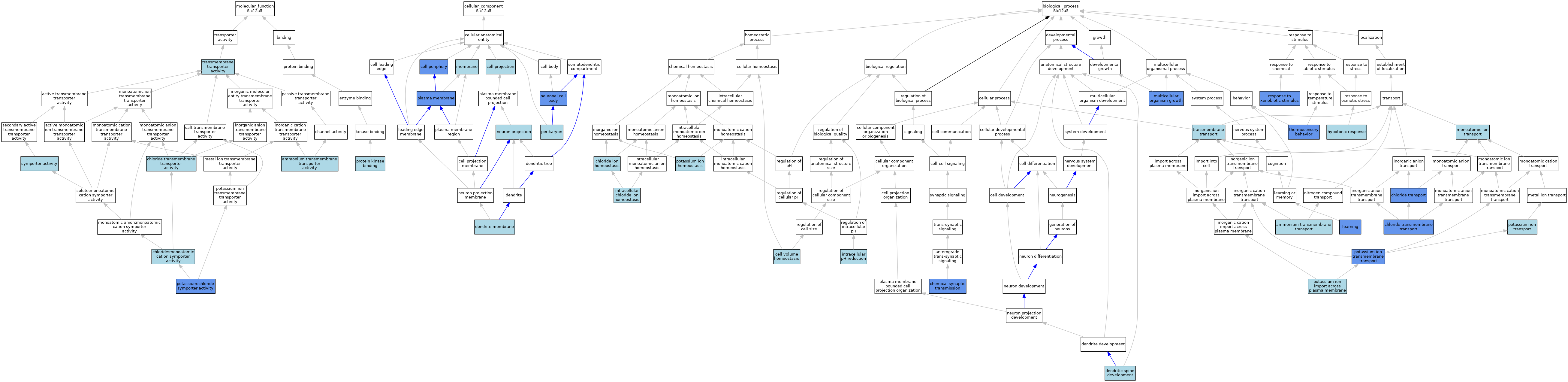
| Molecular Function | GO:0008519 | ammonium transmembrane transporter activity | ISO | J:155856 | |||||||||
| Molecular Function | GO:0015108 | chloride transmembrane transporter activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0015377 | chloride:monoatomic cation symporter activity | IEA | J:72247 | |||||||||
| Molecular Function | GO:0015379 | potassium:chloride symporter activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0015379 | potassium:chloride symporter activity | ISO | J:155856 | |||||||||
| Molecular Function | GO:0015379 | potassium:chloride symporter activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0015379 | potassium:chloride symporter activity | IMP | J:69625 | |||||||||
| Molecular Function | GO:0019901 | protein kinase binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0015293 | symporter activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0022857 | transmembrane transporter activity | IEA | J:72247 | |||||||||
| Cellular Component | GO:0071944 | cell periphery | IDA | J:219851 | |||||||||
| Cellular Component | GO:0071944 | cell periphery | ISO | J:164563 | |||||||||
| Cellular Component | GO:0042995 | cell projection | IEA | J:60000 | |||||||||
| Cellular Component | GO:0032590 | dendrite membrane | ISO | J:155856 | |||||||||
| Cellular Component | GO:0016020 | membrane | ISO | J:164563 | |||||||||
| Cellular Component | GO:0043005 | neuron projection | ISO | J:164563 | |||||||||
| Cellular Component | GO:0043025 | neuronal cell body | ISO | J:164563 | |||||||||
| Cellular Component | GO:0043025 | neuronal cell body | IDA | J:219851 | |||||||||
| Cellular Component | GO:0043204 | perikaryon | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | IDA | J:69625 | |||||||||
| Biological Process | GO:0072488 | ammonium transmembrane transport | ISO | J:155856 | |||||||||
| Biological Process | GO:0006884 | cell volume homeostasis | IBA | J:265628 | |||||||||
| Biological Process | GO:0007268 | chemical synaptic transmission | IBA | J:265628 | |||||||||
| Biological Process | GO:0007268 | chemical synaptic transmission | IMP | J:69625 | |||||||||
| Biological Process | GO:0055064 | chloride ion homeostasis | IBA | J:265628 | |||||||||
| Biological Process | GO:1902476 | chloride transmembrane transport | IMP | J:69625 | |||||||||
| Biological Process | GO:1902476 | chloride transmembrane transport | IBA | J:265628 | |||||||||
| Biological Process | GO:0006821 | chloride transport | ISO | J:155856 | |||||||||
| Biological Process | GO:0006821 | chloride transport | IMP | J:69625 | |||||||||
| Biological Process | GO:0060996 | dendritic spine development | ISO | J:164563 | |||||||||
| Biological Process | GO:0060996 | dendritic spine development | ISO | J:155856 | |||||||||
| Biological Process | GO:0006971 | hypotonic response | ISO | J:164563 | |||||||||
| Biological Process | GO:0030644 | intracellular chloride ion homeostasis | ISO | J:164563 | |||||||||
| Biological Process | GO:0051452 | intracellular pH reduction | ISO | J:155856 | |||||||||
| Biological Process | GO:0007612 | learning | IMP | J:101079 | |||||||||
| Biological Process | GO:0006811 | monoatomic ion transport | ISO | J:164563 | |||||||||
| Biological Process | GO:0035264 | multicellular organism growth | IMP | J:101079 | |||||||||
| Biological Process | GO:0055075 | potassium ion homeostasis | IBA | J:265628 | |||||||||
| Biological Process | GO:1990573 | potassium ion import across plasma membrane | IBA | J:265628 | |||||||||
| Biological Process | GO:0071805 | potassium ion transmembrane transport | IMP | J:69625 | |||||||||
| Biological Process | GO:0071805 | potassium ion transmembrane transport | ISO | J:155856 | |||||||||
| Biological Process | GO:0006813 | potassium ion transport | ISO | J:155856 | |||||||||
| Biological Process | GO:0009410 | response to xenobiotic stimulus | IMP | J:101079 | |||||||||
| Biological Process | GO:0040040 | thermosensory behavior | IMP | J:101079 | |||||||||
| Biological Process | GO:0055085 | transmembrane transport | IEA | J:72247 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/16/2024 MGI 6.23 |

|
|
|
||