|
Symbol Name ID |
Gabarap
gamma-aminobutyric acid receptor associated protein MGI:1861742 |
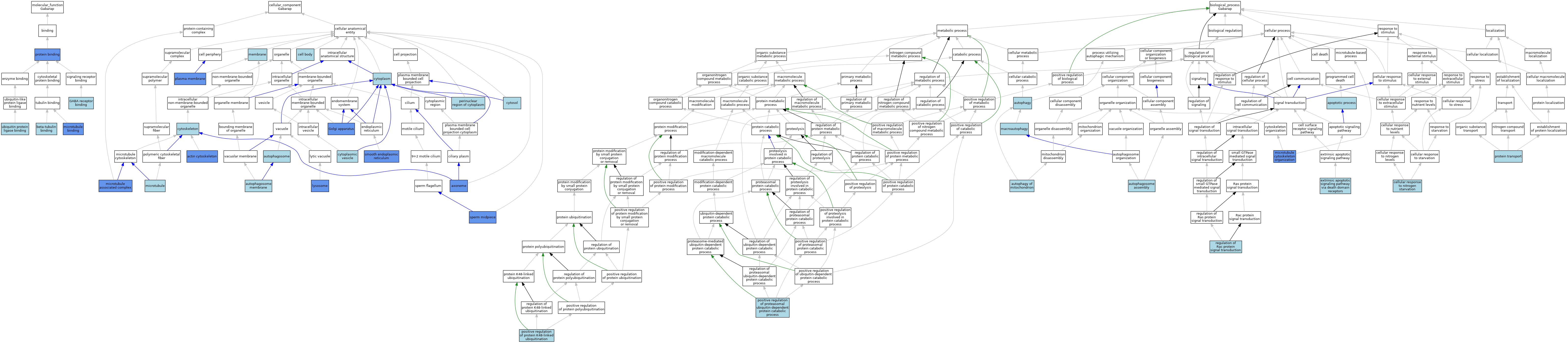
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0048487 | beta-tubulin binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0050811 | GABA receptor binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0050811 | GABA receptor binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0050811 | GABA receptor binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:0008017 | microtubule binding | IDA | J:63439 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:170318 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:63409 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:75687 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:313627 | |||||||||
| Molecular Function | GO:0031625 | ubiquitin protein ligase binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0031625 | ubiquitin protein ligase binding | IBA | J:265628 | |||||||||
| Cellular Component | GO:0015629 | actin cytoskeleton | IDA | J:63439 | |||||||||
| Cellular Component | GO:0005776 | autophagosome | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005776 | autophagosome | IBA | J:265628 | |||||||||
| Cellular Component | GO:0000421 | autophagosome membrane | ISO | J:164563 | |||||||||
| Cellular Component | GO:0000421 | autophagosome membrane | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005930 | axoneme | IDA | J:203423 | |||||||||
| Cellular Component | GO:0044297 | cell body | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IEA | J:60000 | |||||||||
| Cellular Component | GO:0031410 | cytoplasmic vesicle | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005856 | cytoskeleton | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005829 | cytosol | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005794 | Golgi apparatus | IDA | J:67006 | |||||||||
| Cellular Component | GO:0005764 | lysosome | IDA | J:67006 | |||||||||
| Cellular Component | GO:0016020 | membrane | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005874 | microtubule | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005875 | microtubule associated complex | IDA | J:63439 | |||||||||
| Cellular Component | GO:0048471 | perinuclear region of cytoplasm | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | IDA | J:63409 | |||||||||
| Cellular Component | GO:0005790 | smooth endoplasmic reticulum | IDA | J:67006 | |||||||||
| Cellular Component | GO:0097225 | sperm midpiece | IDA | J:178087 | |||||||||
| Biological Process | GO:0006915 | apoptotic process | IEA | J:60000 | |||||||||
| Biological Process | GO:0000045 | autophagosome assembly | IBA | J:265628 | |||||||||
| Biological Process | GO:0006914 | autophagy | IEA | J:60000 | |||||||||
| Biological Process | GO:0000422 | autophagy of mitochondrion | IBA | J:265628 | |||||||||
| Biological Process | GO:0006995 | cellular response to nitrogen starvation | IBA | J:265628 | |||||||||
| Biological Process | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors | ISO | J:164563 | |||||||||
| Biological Process | GO:0016236 | macroautophagy | IBA | J:265628 | |||||||||
| Biological Process | GO:0000226 | microtubule cytoskeleton organization | IDA | J:63439 | |||||||||
| Biological Process | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process | ISO | J:164563 | |||||||||
| Biological Process | GO:1902524 | positive regulation of protein K48-linked ubiquitination | ISO | J:164563 | |||||||||
| Biological Process | GO:0015031 | protein transport | ISO | J:155856 | |||||||||
| Biological Process | GO:0035020 | regulation of Rac protein signal transduction | ISO | J:164563 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/30/2024 MGI 6.23 |

|
|
|
||