|
Symbol Name ID |
Oaz3
ornithine decarboxylase antizyme 3 MGI:1858170 |
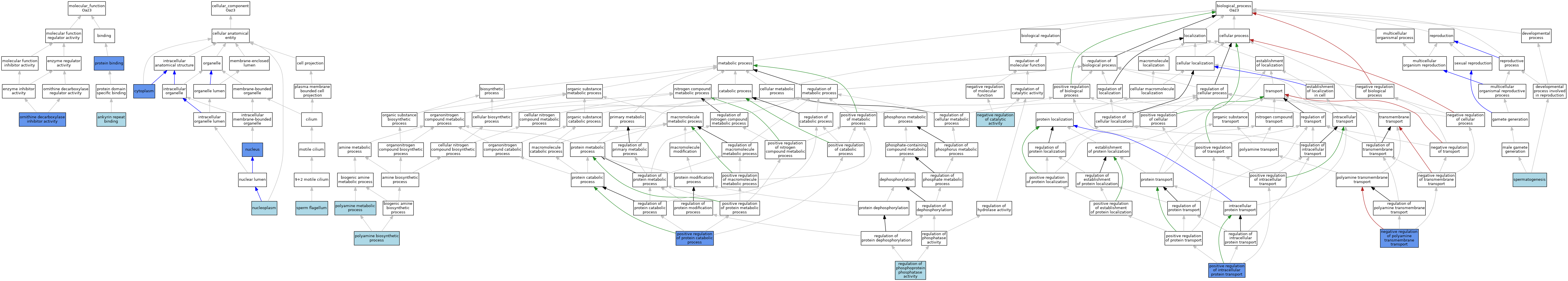
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0071532 | ankyrin repeat binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0008073 | ornithine decarboxylase inhibitor activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0008073 | ornithine decarboxylase inhibitor activity | IDA | J:211327 | |||||||||
| Molecular Function | GO:0008073 | ornithine decarboxylase inhibitor activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0008073 | ornithine decarboxylase inhibitor activity | IGI | J:116975 | |||||||||
| Molecular Function | GO:0008073 | ornithine decarboxylase inhibitor activity | IDA | J:61952 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:213902 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:116975 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:96180 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IDA | J:96180 | |||||||||
| Cellular Component | GO:0005654 | nucleoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IDA | J:96180 | |||||||||
| Cellular Component | GO:0036126 | sperm flagellum | ISO | J:155856 | |||||||||
| Biological Process | GO:0043086 | negative regulation of catalytic activity | IEA | J:72247 | |||||||||
| Biological Process | GO:1902268 | negative regulation of polyamine transmembrane transport | IDA | J:137928 | |||||||||
| Biological Process | GO:1902268 | negative regulation of polyamine transmembrane transport | IDA | J:211327 | |||||||||
| Biological Process | GO:0006596 | polyamine biosynthetic process | IEA | J:60000 | |||||||||
| Biological Process | GO:0006595 | polyamine metabolic process | TAS | J:61952 | |||||||||
| Biological Process | GO:0090316 | positive regulation of intracellular protein transport | IDA | J:212801 | |||||||||
| Biological Process | GO:0045732 | positive regulation of protein catabolic process | IDA | J:211327 | |||||||||
| Biological Process | GO:0045732 | positive regulation of protein catabolic process | IBA | J:265628 | |||||||||
| Biological Process | GO:0043666 | regulation of phosphoprotein phosphatase activity | ISO | J:155856 | |||||||||
| Biological Process | GO:0007283 | spermatogenesis | IEA | J:72247 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/30/2024 MGI 6.23 |

|
|
|
||