|
Symbol Name ID |
Bag3
BCL2-associated athanogene 3 MGI:1352493 |
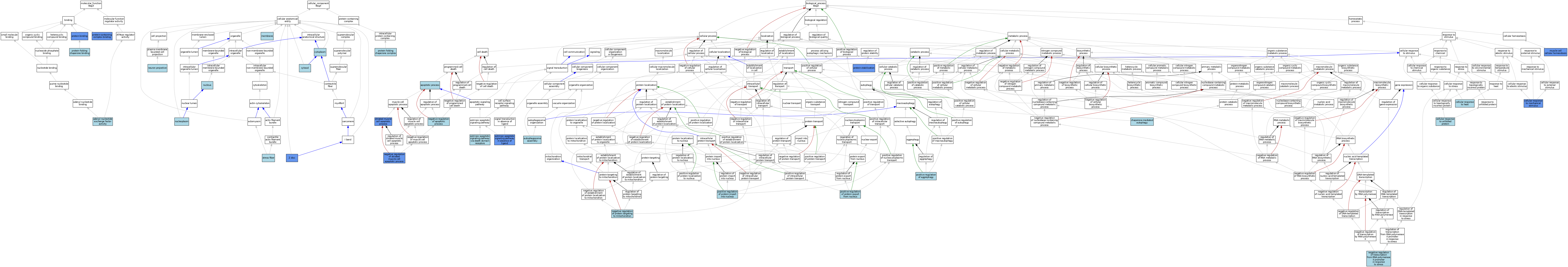
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0000774 | adenyl-nucleotide exchange factor activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0000774 | adenyl-nucleotide exchange factor activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0000774 | adenyl-nucleotide exchange factor activity | ISO | J:156684 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:213186 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:94503 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:156684 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:156684 | |||||||||
| Molecular Function | GO:0044877 | protein-containing complex binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0044877 | protein-containing complex binding | IDA | J:178183 | |||||||||
| Molecular Function | GO:0051087 | protein-folding chaperone binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0051087 | protein-folding chaperone binding | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005829 | cytosol | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005829 | cytosol | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005829 | cytosol | ISO | J:58551 | |||||||||
| Cellular Component | GO:0016020 | membrane | IBA | J:265628 | |||||||||
| Cellular Component | GO:0043005 | neuron projection | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005654 | nucleoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005634 | nucleus | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IBA | J:265628 | |||||||||
| Cellular Component | GO:0101031 | protein folding chaperone complex | ISO | J:164563 | |||||||||
| Cellular Component | GO:0001725 | stress fiber | ISO | J:155856 | |||||||||
| Cellular Component | GO:0030018 | Z disc | IDA | J:112361 | |||||||||
| Cellular Component | GO:0030018 | Z disc | IDA | J:156684 | |||||||||
| Biological Process | GO:0006915 | apoptotic process | IEA | J:60000 | |||||||||
| Biological Process | GO:0000045 | autophagosome assembly | ISO | J:155856 | |||||||||
| Biological Process | GO:0034605 | cellular response to heat | ISO | J:164563 | |||||||||
| Biological Process | GO:0071260 | cellular response to mechanical stimulus | IMP | J:112361 | |||||||||
| Biological Process | GO:0071260 | cellular response to mechanical stimulus | IMP | J:112361 | |||||||||
| Biological Process | GO:0034620 | cellular response to unfolded protein | ISO | J:164563 | |||||||||
| Biological Process | GO:0061684 | chaperone-mediated autophagy | ISO | J:155856 | |||||||||
| Biological Process | GO:0097192 | extrinsic apoptotic signaling pathway in absence of ligand | IDA | J:94503 | |||||||||
| Biological Process | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors | ISO | J:58551 | |||||||||
| Biological Process | GO:0046716 | muscle cell cellular homeostasis | ISO | J:164563 | |||||||||
| Biological Process | GO:0046716 | muscle cell cellular homeostasis | IMP | J:112361 | |||||||||
| Biological Process | GO:0046716 | muscle cell cellular homeostasis | IBA | J:265628 | |||||||||
| Biological Process | GO:0043066 | negative regulation of apoptotic process | ISO | J:155856 | |||||||||
| Biological Process | GO:1903215 | negative regulation of protein targeting to mitochondrion | ISO | J:155856 | |||||||||
| Biological Process | GO:0010664 | negative regulation of striated muscle cell apoptotic process | ISO | J:164563 | |||||||||
| Biological Process | GO:0010664 | negative regulation of striated muscle cell apoptotic process | IBA | J:265628 | |||||||||
| Biological Process | GO:0010664 | negative regulation of striated muscle cell apoptotic process | IMP | J:112361 | |||||||||
| Biological Process | GO:0097201 | negative regulation of transcription from RNA polymerase II promoter in response to stress | ISO | J:164563 | |||||||||
| Biological Process | GO:1905337 | positive regulation of aggrephagy | ISO | J:164563 | |||||||||
| Biological Process | GO:0046827 | positive regulation of protein export from nucleus | ISO | J:164563 | |||||||||
| Biological Process | GO:0042307 | positive regulation of protein import into nucleus | ISO | J:164563 | |||||||||
| Biological Process | GO:0050821 | protein stabilization | ISO | J:164563 | |||||||||
| Biological Process | GO:0050821 | protein stabilization | IBA | J:265628 | |||||||||
| Biological Process | GO:0050821 | protein stabilization | IMP | J:178183 | |||||||||
| Biological Process | GO:0010658 | striated muscle cell apoptotic process | IMP | J:112361 | |||||||||
| Biological Process | GO:0010658 | striated muscle cell apoptotic process | IMP | J:112361 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/16/2024 MGI 6.23 |

|
|
|
||