|
Symbol Name ID |
Map2k6
mitogen-activated protein kinase kinase 6 MGI:1346870 |
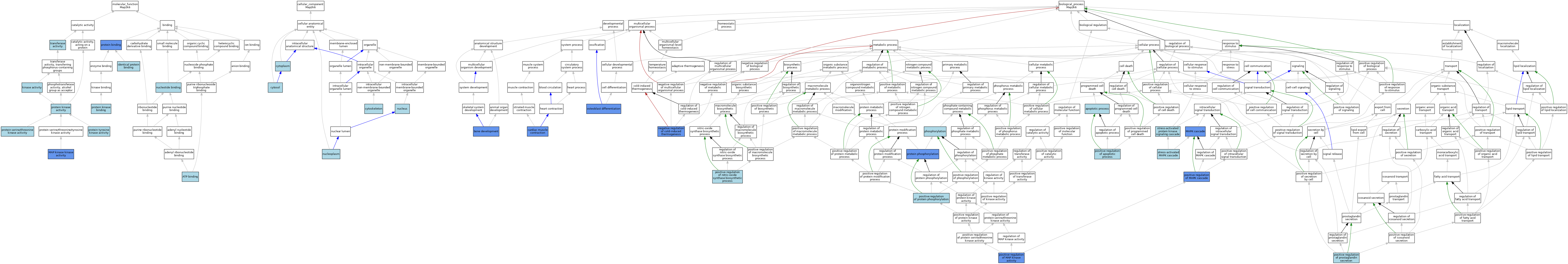
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0005524 | ATP binding | IEA | J:72247 | |||||||||
| Molecular Function | GO:0005524 | ATP binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0042802 | identical protein binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0016301 | kinase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0004708 | MAP kinase kinase activity | ISO | J:155856 | |||||||||
| Molecular Function | GO:0004708 | MAP kinase kinase activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0004708 | MAP kinase kinase activity | TAS | Reactome:R-MMU-448961 | |||||||||
| Molecular Function | GO:0004708 | MAP kinase kinase activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0004708 | MAP kinase kinase activity | IMP | J:163789 | |||||||||
| Molecular Function | GO:0000166 | nucleotide binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:84524 | |||||||||
| Molecular Function | GO:0004672 | protein kinase activity | IEA | J:72247 | |||||||||
| Molecular Function | GO:0019901 | protein kinase binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0004674 | protein serine/threonine kinase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0004713 | protein tyrosine kinase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0016740 | transferase activity | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005856 | cytoskeleton | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005829 | cytosol | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005829 | cytosol | TAS | Reactome:R-MMU-448961 | |||||||||
| Cellular Component | GO:0005829 | cytosol | ISO | J:302013 | |||||||||
| Cellular Component | GO:0005654 | nucleoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IEA | J:60000 | |||||||||
| Biological Process | GO:0006915 | apoptotic process | IEA | J:60000 | |||||||||
| Biological Process | GO:0060348 | bone development | IMP | J:163789 | |||||||||
| Biological Process | GO:0060048 | cardiac muscle contraction | IMP | J:72242 | |||||||||
| Biological Process | GO:0000165 | MAPK cascade | ISO | J:164563 | |||||||||
| Biological Process | GO:0000165 | MAPK cascade | IBA | J:265628 | |||||||||
| Biological Process | GO:0000165 | MAPK cascade | IMP | J:163789 | |||||||||
| Biological Process | GO:0000165 | MAPK cascade | IPI | J:84524 | |||||||||
| Biological Process | GO:0120163 | negative regulation of cold-induced thermogenesis | IMP | J:249909 | |||||||||
| Biological Process | GO:0001649 | osteoblast differentiation | IMP | J:163789 | |||||||||
| Biological Process | GO:0016310 | phosphorylation | IEA | J:60000 | |||||||||
| Biological Process | GO:0043065 | positive regulation of apoptotic process | ISO | J:155856 | |||||||||
| Biological Process | GO:0043406 | positive regulation of MAP kinase activity | IDA | J:88279 | |||||||||
| Biological Process | GO:0043410 | positive regulation of MAPK cascade | IDA | J:97560 | |||||||||
| Biological Process | GO:0043410 | positive regulation of MAPK cascade | IGI | J:97560 | |||||||||
| Biological Process | GO:0043410 | positive regulation of MAPK cascade | IGI | J:97560 | |||||||||
| Biological Process | GO:0043410 | positive regulation of MAPK cascade | IMP | J:72242 | |||||||||
| Biological Process | GO:0051770 | positive regulation of nitric-oxide synthase biosynthetic process | ISO | J:155856 | |||||||||
| Biological Process | GO:0032308 | positive regulation of prostaglandin secretion | ISO | J:155856 | |||||||||
| Biological Process | GO:0001934 | positive regulation of protein phosphorylation | ISO | J:155856 | |||||||||
| Biological Process | GO:0006468 | protein phosphorylation | IDA | J:88279 | |||||||||
| Biological Process | GO:0006468 | protein phosphorylation | IDA | J:79231 | |||||||||
| Biological Process | GO:0051403 | stress-activated MAPK cascade | ISO | J:164563 | |||||||||
| Biological Process | GO:0031098 | stress-activated protein kinase signaling cascade | ISO | J:164563 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/16/2024 MGI 6.23 |

|
|
|
||