|
Symbol Name ID |
Kel
Kell blood group MGI:1346053 |
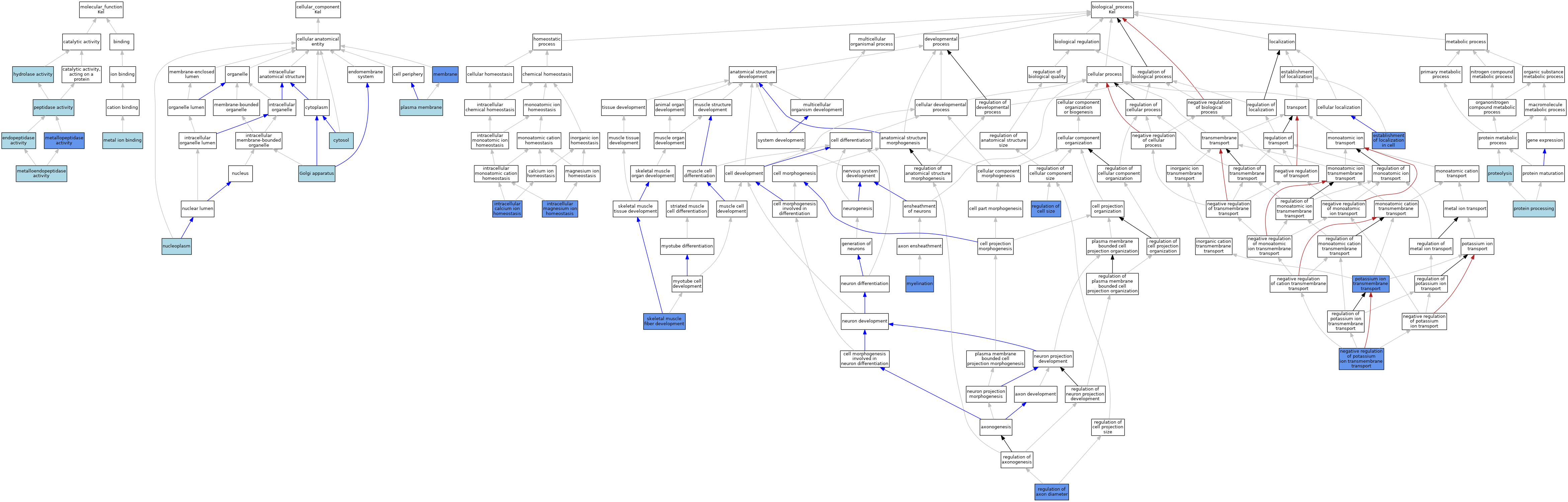
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0004175 | endopeptidase activity | ISS | J:69072 | |||||||||
| Molecular Function | GO:0016787 | hydrolase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0046872 | metal ion binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0004222 | metalloendopeptidase activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0004222 | metalloendopeptidase activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0008237 | metallopeptidase activity | IMP | J:212529 | |||||||||
| Molecular Function | GO:0008233 | peptidase activity | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005829 | cytosol | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005794 | Golgi apparatus | ISO | J:164563 | |||||||||
| Cellular Component | GO:0016020 | membrane | IDA | J:212529 | |||||||||
| Cellular Component | GO:0005654 | nucleoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | IBA | J:265628 | |||||||||
| Biological Process | GO:0051649 | establishment of localization in cell | IGI | J:212529 | |||||||||
| Biological Process | GO:0006874 | intracellular calcium ion homeostasis | IGI | J:212528 | |||||||||
| Biological Process | GO:0006874 | intracellular calcium ion homeostasis | IGI | J:212528 | |||||||||
| Biological Process | GO:0010961 | intracellular magnesium ion homeostasis | IGI | J:212528 | |||||||||
| Biological Process | GO:0010961 | intracellular magnesium ion homeostasis | IGI | J:212528 | |||||||||
| Biological Process | GO:0042552 | myelination | IMP | J:206464 | |||||||||
| Biological Process | GO:0042552 | myelination | IMP | J:206464 | |||||||||
| Biological Process | GO:1901380 | negative regulation of potassium ion transmembrane transport | IGI | J:212529 | |||||||||
| Biological Process | GO:0071805 | potassium ion transmembrane transport | IGI | J:212529 | |||||||||
| Biological Process | GO:0016485 | protein processing | IBA | J:265628 | |||||||||
| Biological Process | GO:0006508 | proteolysis | IEA | J:60000 | |||||||||
| Biological Process | GO:0006508 | proteolysis | IEA | J:72247 | |||||||||
| Biological Process | GO:0031133 | regulation of axon diameter | IGI | J:206464 | |||||||||
| Biological Process | GO:0008361 | regulation of cell size | IGI | J:212528 | |||||||||
| Biological Process | GO:0048741 | skeletal muscle fiber development | IGI | J:206464 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/16/2024 MGI 6.23 |

|
|
|
||