|
Symbol Name ID |
Sh2b2
SH2B adaptor protein 2 MGI:1345171 |
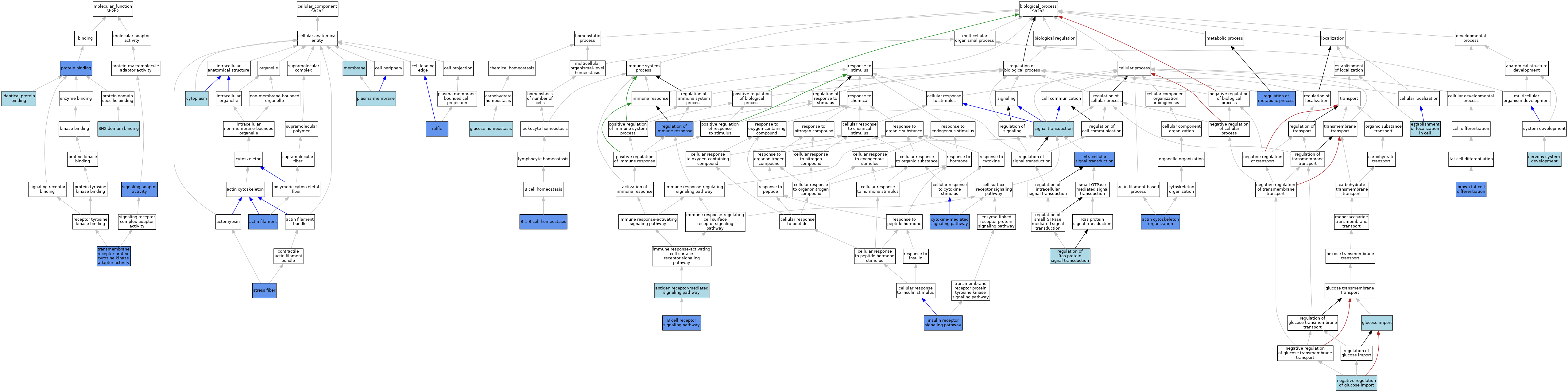
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0042802 | identical protein binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:76351 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:62510 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:100934 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:54187 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:62510 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:62510 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:62510 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:62510 | |||||||||
| Molecular Function | GO:0042169 | SH2 domain binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0035591 | signaling adaptor activity | IDA | J:62510 | |||||||||
| Molecular Function | GO:0035591 | signaling adaptor activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity | IDA | J:54187 | |||||||||
| Cellular Component | GO:0005884 | actin filament | IDA | J:88527 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | ISS | J:113810 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0016020 | membrane | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | ISS | J:113810 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | IBA | J:265628 | |||||||||
| Cellular Component | GO:0001726 | ruffle | IDA | J:100934 | |||||||||
| Cellular Component | GO:0001725 | stress fiber | IDA | J:100934 | |||||||||
| Biological Process | GO:0030036 | actin cytoskeleton organization | IMP | J:88527 | |||||||||
| Biological Process | GO:0030036 | actin cytoskeleton organization | IPI | J:100934 | |||||||||
| Biological Process | GO:0050851 | antigen receptor-mediated signaling pathway | IBA | J:265628 | |||||||||
| Biological Process | GO:0050853 | B cell receptor signaling pathway | ISO | J:164563 | |||||||||
| Biological Process | GO:0050853 | B cell receptor signaling pathway | IDA | J:62510 | |||||||||
| Biological Process | GO:0001922 | B-1 B cell homeostasis | IMP | J:88527 | |||||||||
| Biological Process | GO:0050873 | brown fat cell differentiation | IDA | J:137608 | |||||||||
| Biological Process | GO:0019221 | cytokine-mediated signaling pathway | IDA | J:62510 | |||||||||
| Biological Process | GO:0051649 | establishment of localization in cell | ISA | J:86301 | |||||||||
| Biological Process | GO:0042593 | glucose homeostasis | ISA | J:86301 | |||||||||
| Biological Process | GO:0046323 | glucose import | ISA | J:86301 | |||||||||
| Biological Process | GO:0008286 | insulin receptor signaling pathway | IDA | J:54187 | |||||||||
| Biological Process | GO:0035556 | intracellular signal transduction | IDA | J:62510 | |||||||||
| Biological Process | GO:0035556 | intracellular signal transduction | IBA | J:265628 | |||||||||
| Biological Process | GO:0046325 | negative regulation of glucose import | ISA | J:86301 | |||||||||
| Biological Process | GO:0007399 | nervous system development | ISO | J:155856 | |||||||||
| Biological Process | GO:0050776 | regulation of immune response | IMP | J:88527 | |||||||||
| Biological Process | GO:0019222 | regulation of metabolic process | IMP | J:86301 | |||||||||
| Biological Process | GO:0046578 | regulation of Ras protein signal transduction | ISO | J:155856 | |||||||||
| Biological Process | GO:0007165 | signal transduction | IEA | J:72247 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/30/2024 MGI 6.23 |

|
|
|
||