|
Symbol Name ID |
Rb1cc1
RB1-inducible coiled-coil 1 MGI:1341850 |
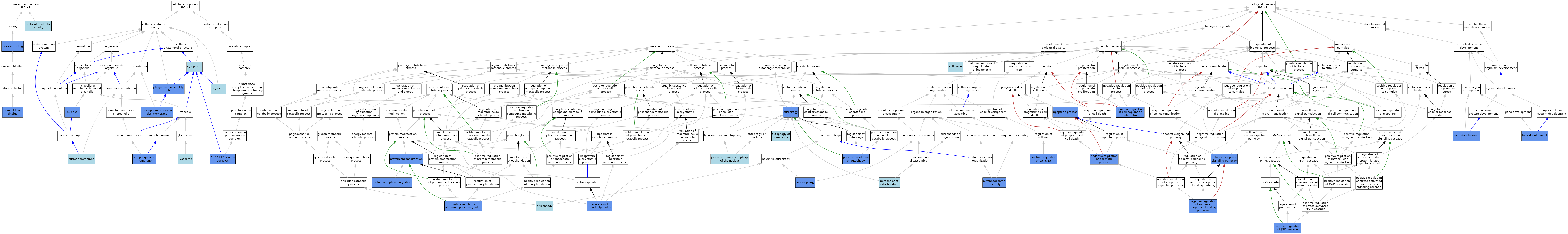
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0060090 | molecular adaptor activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:245290 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:149848 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:281251 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:200458 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:215480 | |||||||||
| Molecular Function | GO:0019901 | protein kinase binding | IPI | J:136659 | |||||||||
| Molecular Function | GO:0019901 | protein kinase binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0019901 | protein kinase binding | IBA | J:265628 | |||||||||
| Cellular Component | GO:1990316 | Atg1/ULK1 kinase complex | ISO | J:164563 | |||||||||
| Cellular Component | GO:1990316 | Atg1/ULK1 kinase complex | IPI | J:149848 | |||||||||
| Cellular Component | GO:1990316 | Atg1/ULK1 kinase complex | IBA | J:265628 | |||||||||
| Cellular Component | GO:1990316 | Atg1/ULK1 kinase complex | IDA | J:153377 | |||||||||
| Cellular Component | GO:0000421 | autophagosome membrane | IDA | J:278382 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | ISS | J:66320 | |||||||||
| Cellular Component | GO:0005829 | cytosol | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005764 | lysosome | ISO | J:164563 | |||||||||
| Cellular Component | GO:0031965 | nuclear membrane | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IDA | J:77935 | |||||||||
| Cellular Component | GO:0005634 | nucleus | ISS | J:66320 | |||||||||
| Cellular Component | GO:0000407 | phagophore assembly site | IDA | J:149848 | |||||||||
| Cellular Component | GO:0000407 | phagophore assembly site | IDA | J:314077 | |||||||||
| Cellular Component | GO:0000407 | phagophore assembly site | ISO | J:164563 | |||||||||
| Cellular Component | GO:0034045 | phagophore assembly site membrane | IDA | J:136659 | |||||||||
| Cellular Component | GO:0034045 | phagophore assembly site membrane | IBA | J:265628 | |||||||||
| Biological Process | GO:0006915 | apoptotic process | IMP | J:114498 | |||||||||
| Biological Process | GO:0006915 | apoptotic process | IMP | J:114498 | |||||||||
| Biological Process | GO:0000045 | autophagosome assembly | ISO | J:164563 | |||||||||
| Biological Process | GO:0000045 | autophagosome assembly | IMP | J:149848 | |||||||||
| Biological Process | GO:0000045 | autophagosome assembly | IDA | J:314078 | |||||||||
| Biological Process | GO:0000045 | autophagosome assembly | IBA | J:265628 | |||||||||
| Biological Process | GO:0006914 | autophagy | IMP | J:136659 | |||||||||
| Biological Process | GO:0006914 | autophagy | IMP | J:278382 | |||||||||
| Biological Process | GO:0000422 | autophagy of mitochondrion | IBA | J:265628 | |||||||||
| Biological Process | GO:0030242 | autophagy of peroxisome | IBA | J:265628 | |||||||||
| Biological Process | GO:0007049 | cell cycle | IEA | J:60000 | |||||||||
| Biological Process | GO:0097191 | extrinsic apoptotic signaling pathway | IMP | J:114498 | |||||||||
| Biological Process | GO:0097191 | extrinsic apoptotic signaling pathway | IMP | J:114498 | |||||||||
| Biological Process | GO:0061723 | glycophagy | IBA | J:265628 | |||||||||
| Biological Process | GO:0007507 | heart development | IMP | J:114498 | |||||||||
| Biological Process | GO:0001889 | liver development | IMP | J:114498 | |||||||||
| Biological Process | GO:0043066 | negative regulation of apoptotic process | IMP | J:114498 | |||||||||
| Biological Process | GO:0008285 | negative regulation of cell population proliferation | IDA | J:314079 | |||||||||
| Biological Process | GO:0008285 | negative regulation of cell population proliferation | ISO | J:164563 | |||||||||
| Biological Process | GO:2001237 | negative regulation of extrinsic apoptotic signaling pathway | IMP | J:114498 | |||||||||
| Biological Process | GO:0034727 | piecemeal microautophagy of the nucleus | IBA | J:265628 | |||||||||
| Biological Process | GO:0010508 | positive regulation of autophagy | IDA | J:314078 | |||||||||
| Biological Process | GO:0045793 | positive regulation of cell size | IMP | J:114498 | |||||||||
| Biological Process | GO:0045793 | positive regulation of cell size | IMP | J:114498 | |||||||||
| Biological Process | GO:0046330 | positive regulation of JNK cascade | IGI | J:114498 | |||||||||
| Biological Process | GO:0001934 | positive regulation of protein phosphorylation | IBA | J:265628 | |||||||||
| Biological Process | GO:0001934 | positive regulation of protein phosphorylation | IMP | J:114498 | |||||||||
| Biological Process | GO:0046777 | protein autophosphorylation | IDA | J:175316 | |||||||||
| Biological Process | GO:0046777 | protein autophosphorylation | ISO | J:164563 | |||||||||
| Biological Process | GO:0006468 | protein phosphorylation | ISO | J:164563 | |||||||||
| Biological Process | GO:0006468 | protein phosphorylation | IDA | J:314078 | |||||||||
| Biological Process | GO:0006468 | protein phosphorylation | IMP | J:114498 | |||||||||
| Biological Process | GO:0006468 | protein phosphorylation | IMP | J:114498 | |||||||||
| Biological Process | GO:0006468 | protein phosphorylation | IMP | J:114498 | |||||||||
| Biological Process | GO:1903059 | regulation of protein lipidation | IDA | J:314078 | |||||||||
| Biological Process | GO:1903059 | regulation of protein lipidation | ISO | J:164563 | |||||||||
| Biological Process | GO:0061709 | reticulophagy | IBA | J:265628 | |||||||||
| Biological Process | GO:0061709 | reticulophagy | IMP | J:278382 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/30/2024 MGI 6.23 |

|
|
|
||