|
Symbol Name ID |
Zfp207
zinc finger protein 207 MGI:1340045 |
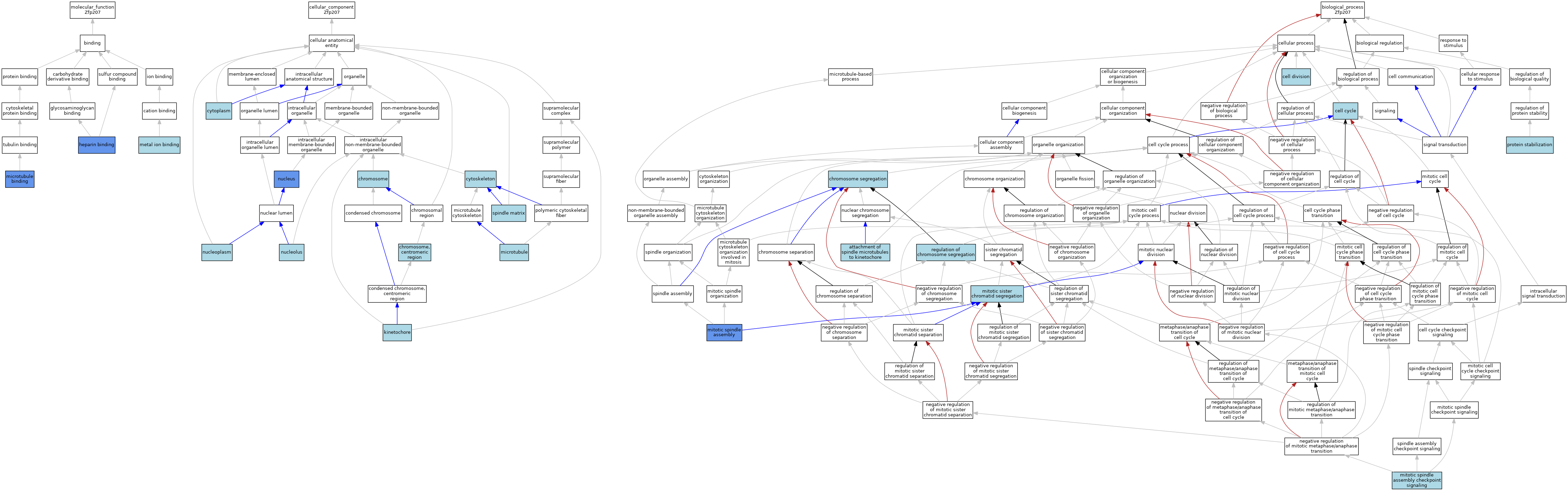
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0008201 | heparin binding | IDA | J:54760 | |||||||||
| Molecular Function | GO:0046872 | metal ion binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0008017 | microtubule binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0008017 | microtubule binding | IDA | J:225576 | |||||||||
| Cellular Component | GO:0005694 | chromosome | IEA | J:60000 | |||||||||
| Cellular Component | GO:0000775 | chromosome, centromeric region | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005856 | cytoskeleton | IEA | J:60000 | |||||||||
| Cellular Component | GO:0000776 | kinetochore | ISO | J:164563 | |||||||||
| Cellular Component | GO:0000776 | kinetochore | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005874 | microtubule | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005730 | nucleolus | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005654 | nucleoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005634 | nucleus | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IDA | J:64959 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IDA | J:54760 | |||||||||
| Cellular Component | GO:1990047 | spindle matrix | ISO | J:164563 | |||||||||
| Cellular Component | GO:1990047 | spindle matrix | IBA | J:265628 | |||||||||
| Biological Process | GO:0008608 | attachment of spindle microtubules to kinetochore | ISO | J:164563 | |||||||||
| Biological Process | GO:0008608 | attachment of spindle microtubules to kinetochore | IBA | J:265628 | |||||||||
| Biological Process | GO:0007049 | cell cycle | IEA | J:60000 | |||||||||
| Biological Process | GO:0051301 | cell division | IEA | J:60000 | |||||||||
| Biological Process | GO:0007059 | chromosome segregation | IEA | J:60000 | |||||||||
| Biological Process | GO:0000070 | mitotic sister chromatid segregation | ISO | J:164563 | |||||||||
| Biological Process | GO:0090307 | mitotic spindle assembly | IDA | J:225576 | |||||||||
| Biological Process | GO:0090307 | mitotic spindle assembly | ISO | J:164563 | |||||||||
| Biological Process | GO:0007094 | mitotic spindle assembly checkpoint signaling | ISO | J:164563 | |||||||||
| Biological Process | GO:0007094 | mitotic spindle assembly checkpoint signaling | IBA | J:265628 | |||||||||
| Biological Process | GO:0050821 | protein stabilization | ISO | J:164563 | |||||||||
| Biological Process | GO:0051983 | regulation of chromosome segregation | ISO | J:164563 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/16/2024 MGI 6.23 |

|
|
|
||