|
Symbol Name ID |
Socs3
suppressor of cytokine signaling 3 MGI:1201791 |
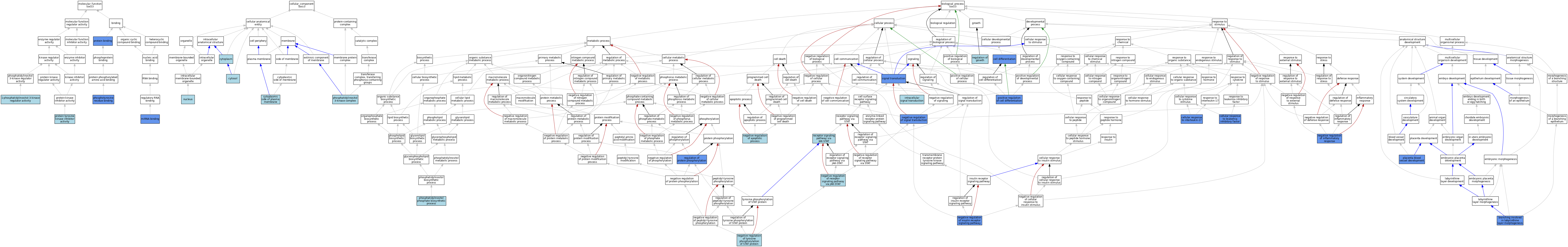
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0035198 | miRNA binding | IDA | J:269278 | |||||||||
| Molecular Function | GO:0035198 | miRNA binding | IMP | J:269278 | |||||||||
| Molecular Function | GO:0001784 | phosphotyrosine residue binding | IMP | J:240281 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:304576 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:168875 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:201263 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:91027 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:94679 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:94679 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:94679 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:94679 | |||||||||
| Molecular Function | GO:0030292 | protein tyrosine kinase inhibitor activity | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | ISO | J:155856 | |||||||||
| Cellular Component | GO:0009898 | cytoplasmic side of plasma membrane | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005829 | cytosol | TAS | Reactome:R-MMU-1169194 | |||||||||
| Cellular Component | GO:0005829 | cytosol | TAS | Reactome:R-MMU-2672295 | |||||||||
| Cellular Component | GO:0005829 | cytosol | TAS | Reactome:R-MMU-8955856 | |||||||||
| Cellular Component | GO:0005829 | cytosol | TAS | Reactome:R-MMU-9705464 | |||||||||
| Cellular Component | GO:0005829 | cytosol | TAS | Reactome:R-MMU-9705471 | |||||||||
| Cellular Component | GO:0005829 | cytosol | TAS | Reactome:R-MMU-9707972 | |||||||||
| Cellular Component | GO:0005829 | cytosol | TAS | Reactome:R-MMU-9707979 | |||||||||
| Cellular Component | GO:0005634 | nucleus | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005942 | phosphatidylinositol 3-kinase complex | IBA | J:265628 | |||||||||
| Biological Process | GO:0060670 | branching involved in labyrinthine layer morphogenesis | IMP | J:70667 | |||||||||
| Biological Process | GO:0030154 | cell differentiation | IGI | J:96515 | |||||||||
| Biological Process | GO:0030154 | cell differentiation | IDA | J:96515 | |||||||||
| Biological Process | GO:0097398 | cellular response to interleukin-17 | IDA | J:269278 | |||||||||
| Biological Process | GO:1990830 | cellular response to leukemia inhibitory factor | IEP | J:163558 | |||||||||
| Biological Process | GO:0035556 | intracellular signal transduction | ISO | J:155856 | |||||||||
| Biological Process | GO:0043066 | negative regulation of apoptotic process | IEA | J:72247 | |||||||||
| Biological Process | GO:0050728 | negative regulation of inflammatory response | IGI | J:163717 | |||||||||
| Biological Process | GO:0046627 | negative regulation of insulin receptor signaling pathway | IDA | J:91027 | |||||||||
| Biological Process | GO:0046426 | negative regulation of receptor signaling pathway via JAK-STAT | ISO | J:164563 | |||||||||
| Biological Process | GO:0009968 | negative regulation of signal transduction | IGI | J:96515 | |||||||||
| Biological Process | GO:0042532 | negative regulation of tyrosine phosphorylation of STAT protein | ISO | J:164563 | |||||||||
| Biological Process | GO:0046854 | phosphatidylinositol phosphate biosynthetic process | IBA | J:265628 | |||||||||
| Biological Process | GO:0060674 | placenta blood vessel development | IMP | J:70667 | |||||||||
| Biological Process | GO:0045597 | positive regulation of cell differentiation | IDA | J:96515 | |||||||||
| Biological Process | GO:0045597 | positive regulation of cell differentiation | IGI | J:96515 | |||||||||
| Biological Process | GO:0007259 | receptor signaling pathway via JAK-STAT | ISO | J:155856 | |||||||||
| Biological Process | GO:0040008 | regulation of growth | IEA | J:60000 | |||||||||
| Biological Process | GO:0001932 | regulation of protein phosphorylation | IDA | J:91027 | |||||||||
| Biological Process | GO:0007165 | signal transduction | ISO | J:155856 | |||||||||
| Biological Process | GO:0007165 | signal transduction | IGI | J:96515 | |||||||||
| Biological Process | GO:0007165 | signal transduction | IGI | J:96515 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/30/2024 MGI 6.23 |

|
|
|
||