|
Symbol Name ID |
Lmx1b
LIM homeobox transcription factor 1 beta MGI:1100513 |
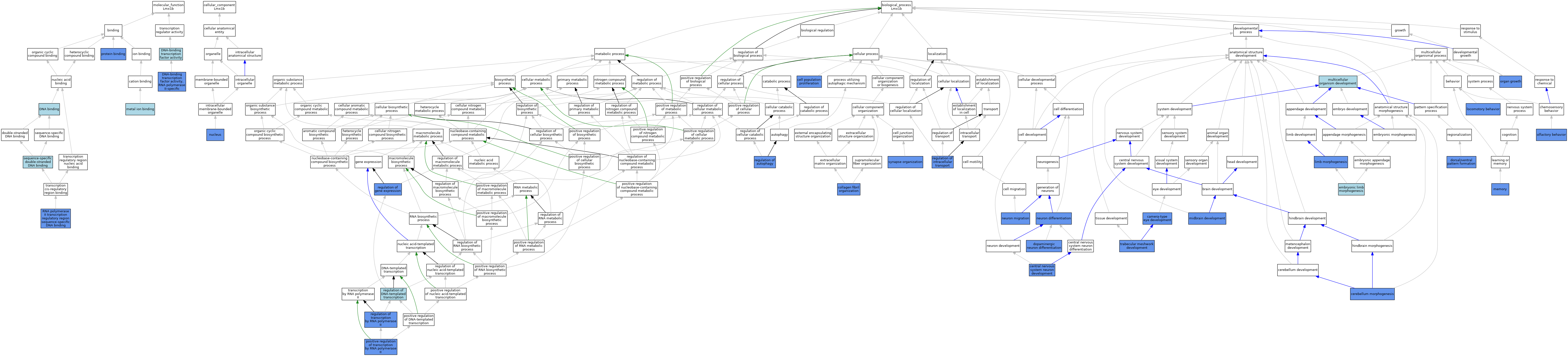
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0003677 | DNA binding | IEA | J:72247 | |||||||||
| Molecular Function | GO:0003677 | DNA binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0003700 | DNA-binding transcription factor activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0000981 | DNA-binding transcription factor activity, RNA polymerase II-specific | IDA | J:228113 | |||||||||
| Molecular Function | GO:0000981 | DNA-binding transcription factor activity, RNA polymerase II-specific | IBA | J:265628 | |||||||||
| Molecular Function | GO:0046872 | metal ion binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:242466 | |||||||||
| Molecular Function | GO:0000977 | RNA polymerase II transcription regulatory region sequence-specific DNA binding | IDA | J:228113 | |||||||||
| Molecular Function | GO:0000977 | RNA polymerase II transcription regulatory region sequence-specific DNA binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:1990837 | sequence-specific double-stranded DNA binding | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005634 | nucleus | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IDA | J:130846 | |||||||||
| Biological Process | GO:0043010 | camera-type eye development | IMP | J:60307 | |||||||||
| Biological Process | GO:0043010 | camera-type eye development | IMP | J:161793 | |||||||||
| Biological Process | GO:0008283 | cell population proliferation | IMP | J:117035 | |||||||||
| Biological Process | GO:0008283 | cell population proliferation | IMP | J:117035 | |||||||||
| Biological Process | GO:0021954 | central nervous system neuron development | IMP | J:116706 | |||||||||
| Biological Process | GO:0021587 | cerebellum morphogenesis | IMP | J:117035 | |||||||||
| Biological Process | GO:0030199 | collagen fibril organization | IMP | J:60307 | |||||||||
| Biological Process | GO:0071542 | dopaminergic neuron differentiation | IGI | J:228113 | |||||||||
| Biological Process | GO:0071542 | dopaminergic neuron differentiation | TAS | J:227390 | |||||||||
| Biological Process | GO:0071542 | dopaminergic neuron differentiation | IMP | J:175535 | |||||||||
| Biological Process | GO:0009953 | dorsal/ventral pattern formation | IEP | J:113891 | |||||||||
| Biological Process | GO:0030326 | embryonic limb morphogenesis | NAS | J:113891 | |||||||||
| Biological Process | GO:0035108 | limb morphogenesis | IMP | J:161785 | |||||||||
| Biological Process | GO:0007626 | locomotory behavior | IGI | J:222854 | |||||||||
| Biological Process | GO:0007613 | memory | IGI | J:222854 | |||||||||
| Biological Process | GO:0030901 | midbrain development | IMP | J:175535 | |||||||||
| Biological Process | GO:0007275 | multicellular organism development | IEA | J:60000 | |||||||||
| Biological Process | GO:0030182 | neuron differentiation | IBA | J:265628 | |||||||||
| Biological Process | GO:0030182 | neuron differentiation | IMP | J:85552 | |||||||||
| Biological Process | GO:0030182 | neuron differentiation | IMP | J:92064 | |||||||||
| Biological Process | GO:0001764 | neuron migration | IMP | J:92064 | |||||||||
| Biological Process | GO:0042048 | olfactory behavior | IGI | J:222854 | |||||||||
| Biological Process | GO:0035265 | organ growth | IMP | J:117035 | |||||||||
| Biological Process | GO:0035265 | organ growth | IMP | J:117035 | |||||||||
| Biological Process | GO:0045944 | positive regulation of transcription by RNA polymerase II | IDA | J:228113 | |||||||||
| Biological Process | GO:0045944 | positive regulation of transcription by RNA polymerase II | IMP | J:222854 | |||||||||
| Biological Process | GO:0045944 | positive regulation of transcription by RNA polymerase II | IMP | J:117035 | |||||||||
| Biological Process | GO:0045944 | positive regulation of transcription by RNA polymerase II | IMP | J:117035 | |||||||||
| Biological Process | GO:0045944 | positive regulation of transcription by RNA polymerase II | IMP | J:117035 | |||||||||
| Biological Process | GO:0010506 | regulation of autophagy | IMP | J:222854 | |||||||||
| Biological Process | GO:0006355 | regulation of DNA-templated transcription | ISO | J:164563 | |||||||||
| Biological Process | GO:0010468 | regulation of gene expression | IMP | J:161785 | |||||||||
| Biological Process | GO:0032386 | regulation of intracellular transport | IMP | J:222854 | |||||||||
| Biological Process | GO:0006357 | regulation of transcription by RNA polymerase II | IBA | J:265628 | |||||||||
| Biological Process | GO:0006357 | regulation of transcription by RNA polymerase II | IDA | J:175535 | |||||||||
| Biological Process | GO:0050808 | synapse organization | IGI | J:222854 | |||||||||
| Biological Process | GO:0002930 | trabecular meshwork development | IMP | J:161793 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/16/2024 MGI 6.23 |

|
|
|
||