|
Symbol Name ID |
Mfhas1
malignant fibrous histiocytoma amplified sequence 1 MGI:1098644 |
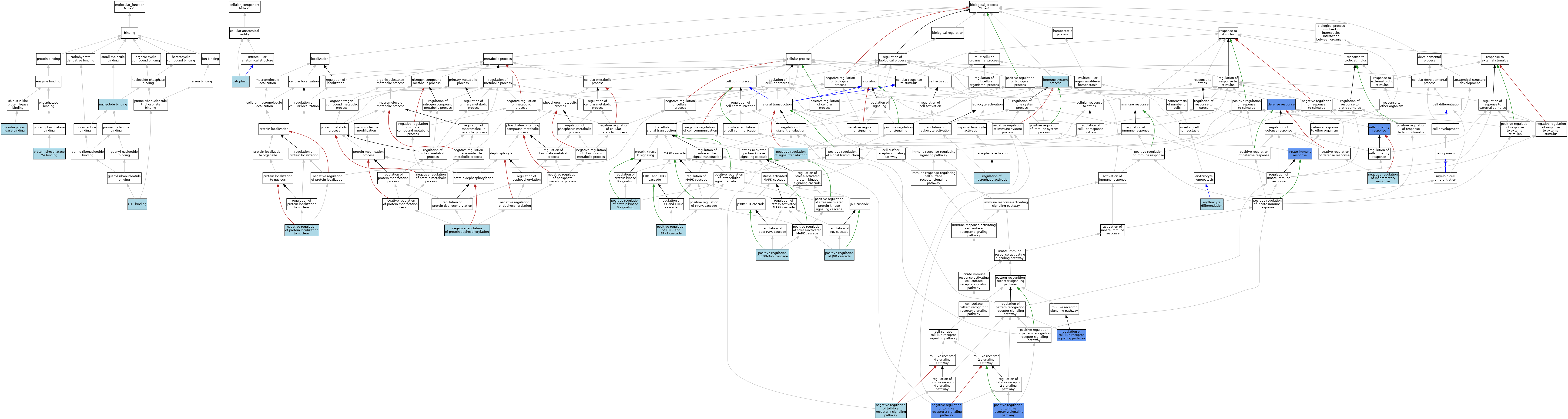
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0005525 | GTP binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0000166 | nucleotide binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0051721 | protein phosphatase 2A binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0031625 | ubiquitin protein ligase binding | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | ISO | J:164563 | |||||||||
| Biological Process | GO:0006952 | defense response | IMP | J:261846 | |||||||||
| Biological Process | GO:0030218 | erythrocyte differentiation | ISO | J:164563 | |||||||||
| Biological Process | GO:0002376 | immune system process | IEA | J:60000 | |||||||||
| Biological Process | GO:0006954 | inflammatory response | ISO | J:164563 | |||||||||
| Biological Process | GO:0006954 | inflammatory response | IMP | J:262346 | |||||||||
| Biological Process | GO:0045087 | innate immune response | IMP | J:262346 | |||||||||
| Biological Process | GO:0045087 | innate immune response | ISO | J:164563 | |||||||||
| Biological Process | GO:0050728 | negative regulation of inflammatory response | ISO | J:164563 | |||||||||
| Biological Process | GO:0035308 | negative regulation of protein dephosphorylation | ISO | J:164563 | |||||||||
| Biological Process | GO:1900181 | negative regulation of protein localization to nucleus | ISO | J:164563 | |||||||||
| Biological Process | GO:0009968 | negative regulation of signal transduction | IEA | J:60000 | |||||||||
| Biological Process | GO:0034136 | negative regulation of toll-like receptor 2 signaling pathway | ISO | J:164563 | |||||||||
| Biological Process | GO:0034136 | negative regulation of toll-like receptor 2 signaling pathway | IMP | J:262346 | |||||||||
| Biological Process | GO:0034144 | negative regulation of toll-like receptor 4 signaling pathway | ISO | J:164563 | |||||||||
| Biological Process | GO:0070374 | positive regulation of ERK1 and ERK2 cascade | ISO | J:164563 | |||||||||
| Biological Process | GO:0046330 | positive regulation of JNK cascade | ISO | J:164563 | |||||||||
| Biological Process | GO:1900745 | positive regulation of p38MAPK cascade | ISO | J:164563 | |||||||||
| Biological Process | GO:0051897 | positive regulation of protein kinase B signaling | ISO | J:164563 | |||||||||
| Biological Process | GO:0034137 | positive regulation of toll-like receptor 2 signaling pathway | ISO | J:164563 | |||||||||
| Biological Process | GO:0034137 | positive regulation of toll-like receptor 2 signaling pathway | IMP | J:262346 | |||||||||
| Biological Process | GO:0043030 | regulation of macrophage activation | ISO | J:164563 | |||||||||
| Biological Process | GO:0034121 | regulation of toll-like receptor signaling pathway | IMP | J:261846 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/30/2024 MGI 6.23 |

|
|
|
||