|
Symbol Name ID |
Ptprq
protein tyrosine phosphatase receptor type Q MGI:1096349 |
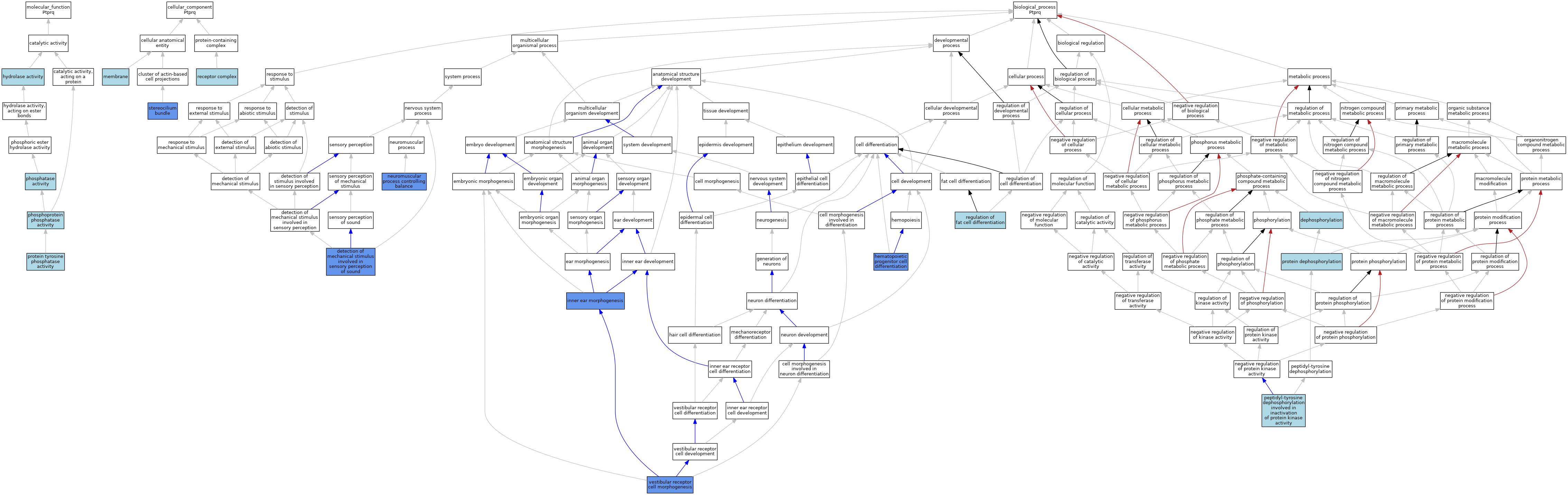
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0016787 | hydrolase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0016791 | phosphatase activity | IEA | J:72247 | |||||||||
| Molecular Function | GO:0004721 | phosphoprotein phosphatase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0004725 | protein tyrosine phosphatase activity | IEA | J:72245 | |||||||||
| Molecular Function | GO:0004725 | protein tyrosine phosphatase activity | IEA | J:72247 | |||||||||
| Cellular Component | GO:0016020 | membrane | IEA | J:60000 | |||||||||
| Cellular Component | GO:0043235 | receptor complex | IBA | J:265628 | |||||||||
| Cellular Component | GO:0032421 | stereocilium bundle | IDA | J:182502 | |||||||||
| Biological Process | GO:0016311 | dephosphorylation | IEA | J:72247 | |||||||||
| Biological Process | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound | IMP | J:182502 | |||||||||
| Biological Process | GO:0002244 | hematopoietic progenitor cell differentiation | IMP | J:202604 | |||||||||
| Biological Process | GO:0042472 | inner ear morphogenesis | IMP | J:85926 | |||||||||
| Biological Process | GO:0050885 | neuromuscular process controlling balance | IMP | J:182502 | |||||||||
| Biological Process | GO:1990264 | peptidyl-tyrosine dephosphorylation involved in inactivation of protein kinase activity | IBA | J:265628 | |||||||||
| Biological Process | GO:0006470 | protein dephosphorylation | IEA | J:72247 | |||||||||
| Biological Process | GO:0045598 | regulation of fat cell differentiation | ISO | J:164563 | |||||||||
| Biological Process | GO:0060116 | vestibular receptor cell morphogenesis | IMP | J:182502 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 05/07/2024 MGI 6.23 |

|
|
|
||