|
Symbol Name ID |
Commd1
COMM domain containing 1 MGI:109474 |
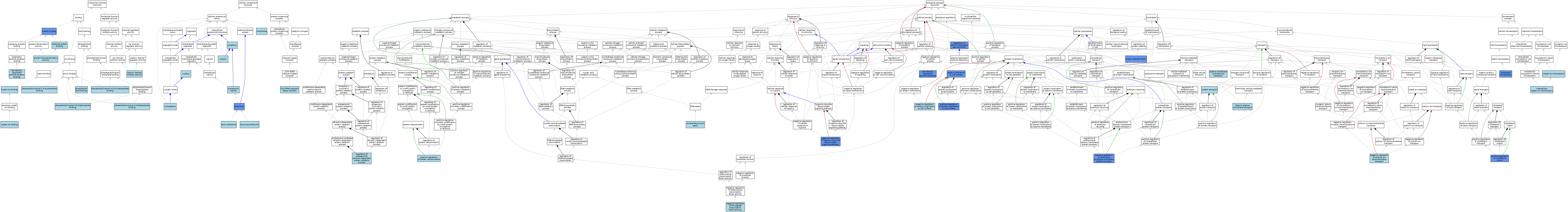
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0005507 | copper ion binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0042802 | identical protein binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0050750 | low-density lipoprotein particle receptor binding | ISO | J:236595 | |||||||||
| Molecular Function | GO:0046872 | metal ion binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0070300 | phosphatidic acid binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:122208 | |||||||||
| Molecular Function | GO:0042803 | protein homodimerization activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0019871 | sodium channel inhibitor activity | ISO | J:73065 | |||||||||
| Cellular Component | GO:0031462 | Cul2-RING ubiquitin ligase complex | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0031410 | cytoplasmic vesicle | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005829 | cytosol | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005769 | early endosome | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005768 | endosome | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005768 | endosome | IDA | J:236595 | |||||||||
| Cellular Component | GO:0016020 | membrane | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005654 | nucleoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005634 | nucleus | ISO | J:164563 | |||||||||
| Cellular Component | GO:0055037 | recycling endosome | ISO | J:164563 | |||||||||
| Biological Process | GO:0042632 | cholesterol homeostasis | ISO | J:236595 | |||||||||
| Biological Process | GO:0042632 | cholesterol homeostasis | IMP | J:236595 | |||||||||
| Biological Process | GO:0055070 | copper ion homeostasis | ISO | J:164563 | |||||||||
| Biological Process | GO:0055070 | copper ion homeostasis | IBA | J:265628 | |||||||||
| Biological Process | GO:0055070 | copper ion homeostasis | ISO | J:236595 | |||||||||
| Biological Process | GO:0006893 | Golgi to plasma membrane transport | ISO | J:164563 | |||||||||
| Biological Process | GO:0006878 | intracellular copper ion homeostasis | ISO | J:74624 | |||||||||
| Biological Process | GO:0034383 | low-density lipoprotein particle clearance | IMP | J:236595 | |||||||||
| Biological Process | GO:1902072 | negative regulation of hypoxia-inducible factor-1alpha signaling pathway | IMP | J:122208 | |||||||||
| Biological Process | GO:0032088 | negative regulation of NF-kappaB transcription factor activity | ISO | J:164563 | |||||||||
| Biological Process | GO:0032088 | negative regulation of NF-kappaB transcription factor activity | ISO | J:115897 | |||||||||
| Biological Process | GO:2000009 | negative regulation of protein localization to cell surface | ISO | J:164563 | |||||||||
| Biological Process | GO:2000009 | negative regulation of protein localization to cell surface | IBA | J:265628 | |||||||||
| Biological Process | GO:1902306 | negative regulation of sodium ion transmembrane transport | ISO | J:164563 | |||||||||
| Biological Process | GO:1902306 | negative regulation of sodium ion transmembrane transport | IBA | J:265628 | |||||||||
| Biological Process | GO:0006289 | nucleotide-excision repair | ISO | J:164563 | |||||||||
| Biological Process | GO:0048227 | plasma membrane to endosome transport | ISO | J:164563 | |||||||||
| Biological Process | GO:1904109 | positive regulation of cholesterol import | IMP | J:236595 | |||||||||
| Biological Process | GO:1905751 | positive regulation of endosome to plasma membrane protein transport | IMP | J:236595 | |||||||||
| Biological Process | GO:2000010 | positive regulation of protein localization to cell surface | IMP | J:236595 | |||||||||
| Biological Process | GO:0031398 | positive regulation of protein ubiquitination | ISO | J:164563 | |||||||||
| Biological Process | GO:0031398 | positive regulation of protein ubiquitination | IBA | J:265628 | |||||||||
| Biological Process | GO:0031648 | protein destabilization | IMP | J:122208 | |||||||||
| Biological Process | GO:0034394 | protein localization to cell surface | IMP | J:236595 | |||||||||
| Biological Process | GO:0015031 | protein transport | IEA | J:60000 | |||||||||
| Biological Process | GO:0097006 | regulation of plasma lipoprotein particle levels | IMP | J:236595 | |||||||||
| Biological Process | GO:0032434 | regulation of proteasomal ubiquitin-dependent protein catabolic process | ISO | J:164563 | |||||||||
| Biological Process | GO:0032434 | regulation of proteasomal ubiquitin-dependent protein catabolic process | IBA | J:265628 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/16/2024 MGI 6.23 |

|
|
|
||