|
Symbol Name ID |
Mapkapk2
MAP kinase-activated protein kinase 2 MGI:109298 |
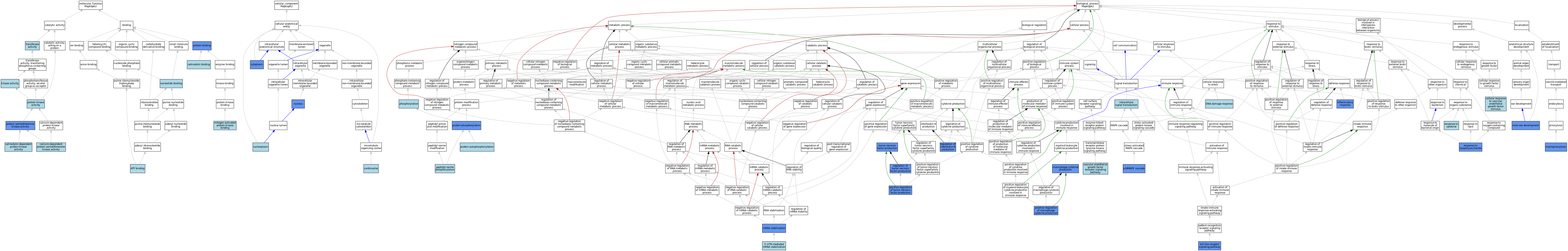
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0005524 | ATP binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0005524 | ATP binding | IEA | J:72247 | |||||||||
| Molecular Function | GO:0009931 | calcium-dependent protein serine/threonine kinase activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0005516 | calmodulin binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:0004683 | calmodulin-dependent protein kinase activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0016301 | kinase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0051019 | mitogen-activated protein kinase binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:0000166 | nucleotide binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:148784 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:126340 | |||||||||
| Molecular Function | GO:0004672 | protein kinase activity | IEA | J:72247 | |||||||||
| Molecular Function | GO:0004674 | protein serine/threonine kinase activity | IDA | J:180488 | |||||||||
| Molecular Function | GO:0004674 | protein serine/threonine kinase activity | IDA | J:180499 | |||||||||
| Molecular Function | GO:0004674 | protein serine/threonine kinase activity | IDA | J:126340 | |||||||||
| Molecular Function | GO:0004674 | protein serine/threonine kinase activity | ISO | J:73065 | |||||||||
| Molecular Function | GO:0004674 | protein serine/threonine kinase activity | IDA | J:204512 | |||||||||
| Molecular Function | GO:0016740 | transferase activity | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005813 | centrosome | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IDA | J:180499 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IDA | J:94780 | |||||||||
| Cellular Component | GO:0005654 | nucleoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IDA | J:180499 | |||||||||
| Cellular Component | GO:0005634 | nucleus | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IDA | J:94780 | |||||||||
| Biological Process | GO:0070935 | 3'-UTR-mediated mRNA stabilization | ISO | J:164563 | |||||||||
| Biological Process | GO:0035924 | cellular response to vascular endothelial growth factor stimulus | ISO | J:164563 | |||||||||
| Biological Process | GO:0035924 | cellular response to vascular endothelial growth factor stimulus | IBA | J:265628 | |||||||||
| Biological Process | GO:0006974 | DNA damage response | ISO | J:164563 | |||||||||
| Biological Process | GO:0006954 | inflammatory response | IMP | J:59678 | |||||||||
| Biological Process | GO:0006954 | inflammatory response | IBA | J:265628 | |||||||||
| Biological Process | GO:0048839 | inner ear development | IDA | J:174607 | |||||||||
| Biological Process | GO:0035556 | intracellular signal transduction | IBA | J:265628 | |||||||||
| Biological Process | GO:0010934 | macrophage cytokine production | IMP | J:204512 | |||||||||
| Biological Process | GO:0044351 | macropinocytosis | IMP | J:126340 | |||||||||
| Biological Process | GO:0048255 | mRNA stabilization | IMP | J:74321 | |||||||||
| Biological Process | GO:0038066 | p38MAPK cascade | IMP | J:204512 | |||||||||
| Biological Process | GO:0018105 | peptidyl-serine phosphorylation | ISO | J:164563 | |||||||||
| Biological Process | GO:0018105 | peptidyl-serine phosphorylation | IBA | J:265628 | |||||||||
| Biological Process | GO:0016310 | phosphorylation | IEA | J:60000 | |||||||||
| Biological Process | GO:0060907 | positive regulation of macrophage cytokine production | IMP | J:204512 | |||||||||
| Biological Process | GO:0032760 | positive regulation of tumor necrosis factor production | IMP | J:204512 | |||||||||
| Biological Process | GO:0046777 | protein autophosphorylation | IBA | J:265628 | |||||||||
| Biological Process | GO:0006468 | protein phosphorylation | IDA | J:180499 | |||||||||
| Biological Process | GO:0032675 | regulation of interleukin-6 production | IMP | J:59678 | |||||||||
| Biological Process | GO:0032680 | regulation of tumor necrosis factor production | ISO | J:164563 | |||||||||
| Biological Process | GO:0032680 | regulation of tumor necrosis factor production | IMP | J:59678 | |||||||||
| Biological Process | GO:0034097 | response to cytokine | ISO | J:164563 | |||||||||
| Biological Process | GO:0034097 | response to cytokine | IBA | J:265628 | |||||||||
| Biological Process | GO:0032496 | response to lipopolysaccharide | IMP | J:59678 | |||||||||
| Biological Process | GO:0032496 | response to lipopolysaccharide | IMP | J:126340 | |||||||||
| Biological Process | GO:0002224 | toll-like receptor signaling pathway | IMP | J:126340 | |||||||||
| Biological Process | GO:0002224 | toll-like receptor signaling pathway | IBA | J:265628 | |||||||||
| Biological Process | GO:0032640 | tumor necrosis factor production | IMP | J:204512 | |||||||||
| Biological Process | GO:0048010 | vascular endothelial growth factor receptor signaling pathway | ISO | J:164563 | |||||||||
| Biological Process | GO:0048010 | vascular endothelial growth factor receptor signaling pathway | IBA | J:265628 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/16/2024 MGI 6.23 |

|
|
|
||