|
Symbol Name ID |
Tubb3
tubulin, beta 3 class III MGI:107813 |
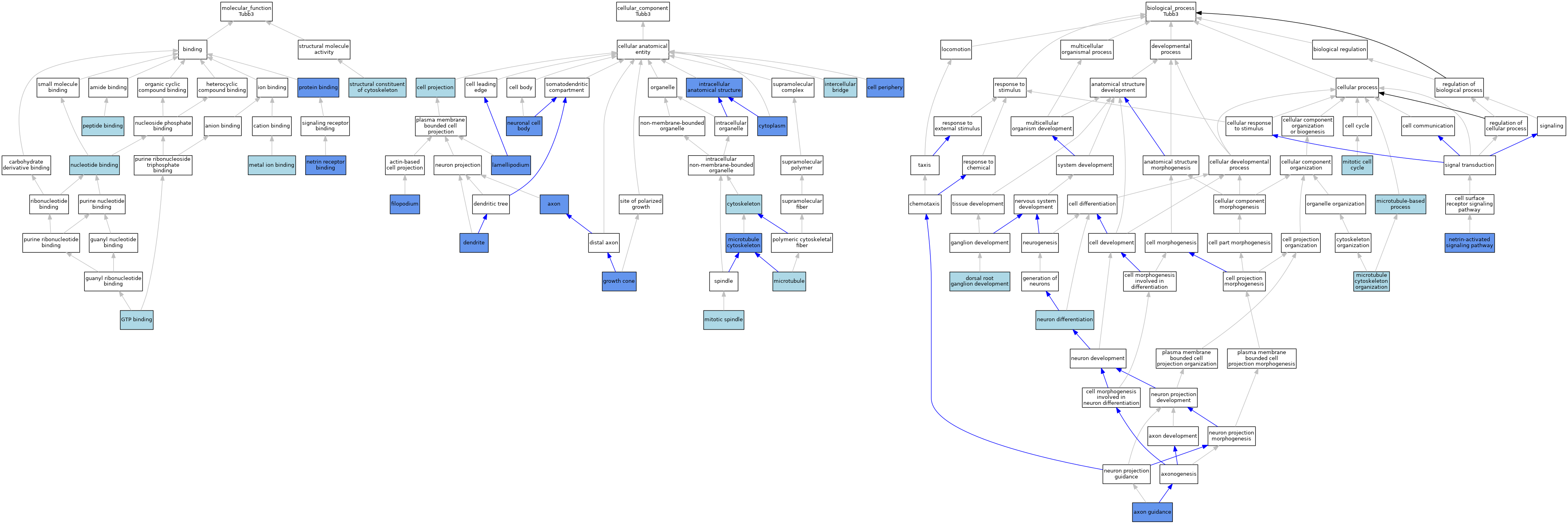
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0005525 | GTP binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0005525 | GTP binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:0046872 | metal ion binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:1990890 | netrin receptor binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:1990890 | netrin receptor binding | IPI | J:242688 | |||||||||
| Molecular Function | GO:0000166 | nucleotide binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0042277 | peptide binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:186067 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:242688 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:154619 | |||||||||
| Molecular Function | GO:0005200 | structural constituent of cytoskeleton | ISO | J:164563 | |||||||||
| Molecular Function | GO:0005200 | structural constituent of cytoskeleton | IBA | J:265628 | |||||||||
| Cellular Component | GO:0030424 | axon | IDA | J:259112 | |||||||||
| Cellular Component | GO:0030424 | axon | IDA | J:130846 | |||||||||
| Cellular Component | GO:0030424 | axon | IDA | J:125222 | |||||||||
| Cellular Component | GO:0030424 | axon | IDA | J:190157 | |||||||||
| Cellular Component | GO:0030424 | axon | IDA | J:139991 | |||||||||
| Cellular Component | GO:0030424 | axon | IDA | J:197573 | |||||||||
| Cellular Component | GO:0030424 | axon | IDA | J:92350 | |||||||||
| Cellular Component | GO:0030424 | axon | ISO | J:156242 | |||||||||
| Cellular Component | GO:0071944 | cell periphery | IDA | J:206879 | |||||||||
| Cellular Component | GO:0042995 | cell projection | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IDA | J:191236 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IDA | J:218609 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IDA | J:190157 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IDA | J:149456 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IDA | J:142500 | |||||||||
| Cellular Component | GO:0005856 | cytoskeleton | IEA | J:60000 | |||||||||
| Cellular Component | GO:0030425 | dendrite | IDA | J:206832 | |||||||||
| Cellular Component | GO:0030175 | filopodium | IDA | J:242688 | |||||||||
| Cellular Component | GO:0030426 | growth cone | IDA | J:242688 | |||||||||
| Cellular Component | GO:0045171 | intercellular bridge | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005622 | intracellular anatomical structure | IDA | J:113146 | |||||||||
| Cellular Component | GO:0030027 | lamellipodium | IDA | J:242688 | |||||||||
| Cellular Component | GO:0005874 | microtubule | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005874 | microtubule | IBA | J:265628 | |||||||||
| Cellular Component | GO:0015630 | microtubule cytoskeleton | ISO | J:164563 | |||||||||
| Cellular Component | GO:0015630 | microtubule cytoskeleton | IDA | J:88813 | |||||||||
| Cellular Component | GO:0072686 | mitotic spindle | ISO | J:164563 | |||||||||
| Cellular Component | GO:0043025 | neuronal cell body | IDA | J:195630 | |||||||||
| Biological Process | GO:0007411 | axon guidance | IMP | J:158992 | |||||||||
| Biological Process | GO:0007411 | axon guidance | ISO | J:164563 | |||||||||
| Biological Process | GO:0007411 | axon guidance | IMP | J:242688 | |||||||||
| Biological Process | GO:0007411 | axon guidance | IBA | J:265628 | |||||||||
| Biological Process | GO:1990791 | dorsal root ganglion development | ISO | J:164563 | |||||||||
| Biological Process | GO:0000226 | microtubule cytoskeleton organization | ISO | J:164563 | |||||||||
| Biological Process | GO:0000226 | microtubule cytoskeleton organization | IBA | J:265628 | |||||||||
| Biological Process | GO:0007017 | microtubule-based process | IEA | J:72247 | |||||||||
| Biological Process | GO:0000278 | mitotic cell cycle | ISO | J:155856 | |||||||||
| Biological Process | GO:0000278 | mitotic cell cycle | IBA | J:265628 | |||||||||
| Biological Process | GO:0038007 | netrin-activated signaling pathway | IMP | J:242688 | |||||||||
| Biological Process | GO:0030182 | neuron differentiation | ISO | J:155856 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 05/07/2024 MGI 6.23 |

|
|
|
||