|
Symbol Name ID |
Trim21
tripartite motif-containing 21 MGI:106657 |
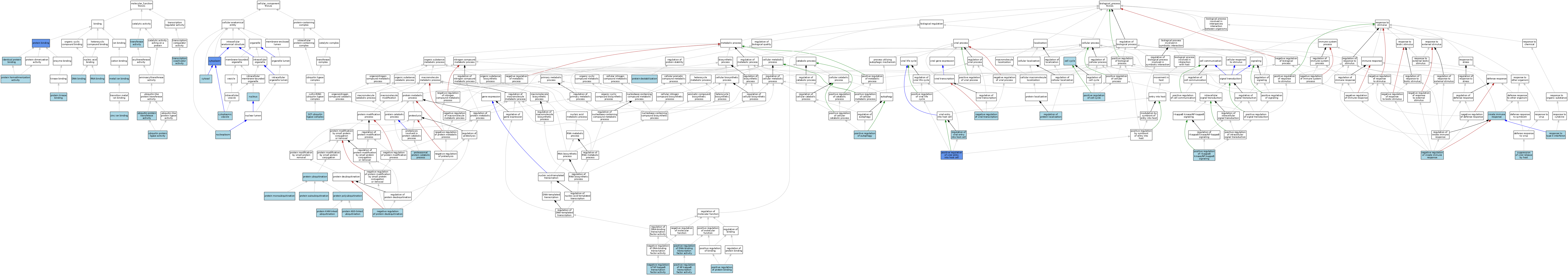
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0003677 | DNA binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0042802 | identical protein binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0046872 | metal ion binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0046872 | metal ion binding | IEA | J:72247 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:192552 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:233627 | |||||||||
| Molecular Function | GO:0042803 | protein homodimerization activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0019901 | protein kinase binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:0003723 | RNA binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0003713 | transcription coactivator activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0016740 | transferase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0061630 | ubiquitin protein ligase activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0061630 | ubiquitin protein ligase activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0004842 | ubiquitin-protein transferase activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0008270 | zinc ion binding | IEA | J:72247 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IDA | J:69273 | |||||||||
| Cellular Component | GO:0031410 | cytoplasmic vesicle | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005829 | cytosol | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005654 | nucleoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005654 | nucleoplasm | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005634 | nucleus | ISO | J:164563 | |||||||||
| Cellular Component | GO:0019005 | SCF ubiquitin ligase complex | ISO | J:164563 | |||||||||
| Biological Process | GO:0007049 | cell cycle | IEA | J:60000 | |||||||||
| Biological Process | GO:0045087 | innate immune response | ISO | J:164563 | |||||||||
| Biological Process | GO:0045087 | innate immune response | IBA | J:265628 | |||||||||
| Biological Process | GO:0045824 | negative regulation of innate immune response | ISO | J:164563 | |||||||||
| Biological Process | GO:0032088 | negative regulation of NF-kappaB transcription factor activity | ISO | J:164563 | |||||||||
| Biological Process | GO:0090086 | negative regulation of protein deubiquitination | ISO | J:164563 | |||||||||
| Biological Process | GO:0032897 | negative regulation of viral transcription | ISO | J:164563 | |||||||||
| Biological Process | GO:0010508 | positive regulation of autophagy | ISO | J:164563 | |||||||||
| Biological Process | GO:0010508 | positive regulation of autophagy | IBA | J:265628 | |||||||||
| Biological Process | GO:0045787 | positive regulation of cell cycle | ISO | J:164563 | |||||||||
| Biological Process | GO:0051091 | positive regulation of DNA-binding transcription factor activity | ISO | J:164563 | |||||||||
| Biological Process | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling | IBA | J:265628 | |||||||||
| Biological Process | GO:0051092 | positive regulation of NF-kappaB transcription factor activity | IBA | J:265628 | |||||||||
| Biological Process | GO:0032092 | positive regulation of protein binding | ISO | J:164563 | |||||||||
| Biological Process | GO:0046598 | positive regulation of viral entry into host cell | IDA | J:205675 | |||||||||
| Biological Process | GO:0010498 | proteasomal protein catabolic process | ISO | J:164563 | |||||||||
| Biological Process | GO:0051865 | protein autoubiquitination | ISO | J:164563 | |||||||||
| Biological Process | GO:0031648 | protein destabilization | ISO | J:164563 | |||||||||
| Biological Process | GO:0070936 | protein K48-linked ubiquitination | ISO | J:164563 | |||||||||
| Biological Process | GO:0070534 | protein K63-linked ubiquitination | ISO | J:164563 | |||||||||
| Biological Process | GO:0006513 | protein monoubiquitination | ISO | J:164563 | |||||||||
| Biological Process | GO:0000209 | protein polyubiquitination | ISO | J:164563 | |||||||||
| Biological Process | GO:0000209 | protein polyubiquitination | IBA | J:265628 | |||||||||
| Biological Process | GO:0016567 | protein ubiquitination | ISO | J:164563 | |||||||||
| Biological Process | GO:0032880 | regulation of protein localization | IBA | J:265628 | |||||||||
| Biological Process | GO:0046596 | regulation of viral entry into host cell | IBA | J:265628 | |||||||||
| Biological Process | GO:0034341 | response to type II interferon | ISO | J:164563 | |||||||||
| Biological Process | GO:0044790 | suppression of viral release by host | ISO | J:164563 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/16/2024 MGI 6.23 |

|
|
|
||