|
Symbol Name ID |
Myo9b
myosin IXb MGI:106624 |
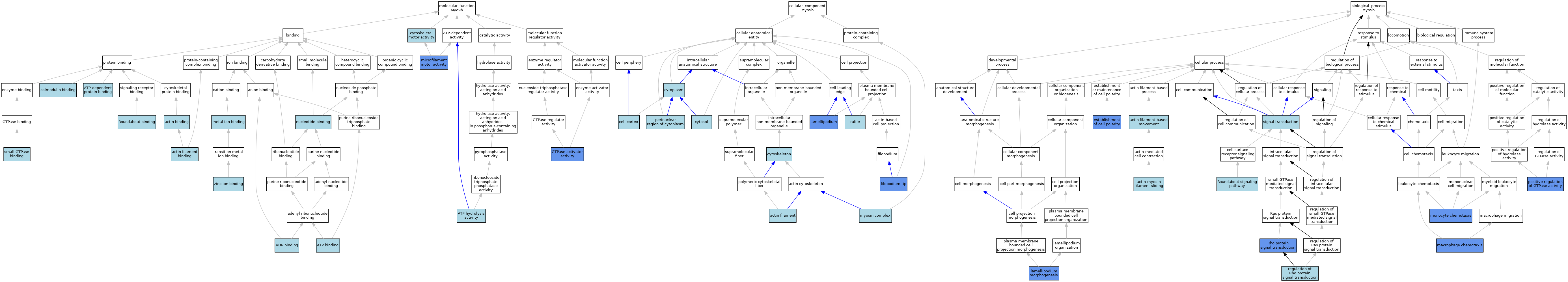
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0003779 | actin binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0003779 | actin binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0003779 | actin binding | ISO | J:73065 | |||||||||
| Molecular Function | GO:0051015 | actin filament binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0051015 | actin filament binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:0051015 | actin filament binding | ISO | J:73065 | |||||||||
| Molecular Function | GO:0043531 | ADP binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0043531 | ADP binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0005524 | ATP binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0005524 | ATP binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0005524 | ATP binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:0005524 | ATP binding | ISO | J:73065 | |||||||||
| Molecular Function | GO:0016887 | ATP hydrolysis activity | ISO | J:155856 | |||||||||
| Molecular Function | GO:0016887 | ATP hydrolysis activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0043008 | ATP-dependent protein binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0005516 | calmodulin binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0003774 | cytoskeletal motor activity | ISO | J:73065 | |||||||||
| Molecular Function | GO:0005096 | GTPase activator activity | ISO | J:155856 | |||||||||
| Molecular Function | GO:0005096 | GTPase activator activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0005096 | GTPase activator activity | IMP | J:162326 | |||||||||
| Molecular Function | GO:0005096 | GTPase activator activity | ISO | J:73065 | |||||||||
| Molecular Function | GO:0005096 | GTPase activator activity | IDA | J:124018 | |||||||||
| Molecular Function | GO:0046872 | metal ion binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0000146 | microfilament motor activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0000146 | microfilament motor activity | ISO | J:155856 | |||||||||
| Molecular Function | GO:0000146 | microfilament motor activity | IDA | J:124018 | |||||||||
| Molecular Function | GO:0000146 | microfilament motor activity | ISO | J:73065 | |||||||||
| Molecular Function | GO:0000166 | nucleotide binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0048495 | Roundabout binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0031267 | small GTPase binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0008270 | zinc ion binding | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005884 | actin filament | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005884 | actin filament | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005938 | cell cortex | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | ISO | J:73065 | |||||||||
| Cellular Component | GO:0005856 | cytoskeleton | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005829 | cytosol | ISO | J:164563 | |||||||||
| Cellular Component | GO:0032433 | filopodium tip | IDA | J:124018 | |||||||||
| Cellular Component | GO:0030027 | lamellipodium | ISO | J:155856 | |||||||||
| Cellular Component | GO:0030027 | lamellipodium | IBA | J:265628 | |||||||||
| Cellular Component | GO:0030027 | lamellipodium | IDA | J:124018 | |||||||||
| Cellular Component | GO:0016459 | myosin complex | IEA | J:60000 | |||||||||
| Cellular Component | GO:0016459 | myosin complex | IEA | J:72247 | |||||||||
| Cellular Component | GO:0048471 | perinuclear region of cytoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0048471 | perinuclear region of cytoplasm | ISO | J:155856 | |||||||||
| Cellular Component | GO:0001726 | ruffle | ISO | J:155856 | |||||||||
| Cellular Component | GO:0001726 | ruffle | IBA | J:265628 | |||||||||
| Biological Process | GO:0030048 | actin filament-based movement | ISO | J:164563 | |||||||||
| Biological Process | GO:0030048 | actin filament-based movement | IBA | J:265628 | |||||||||
| Biological Process | GO:0033275 | actin-myosin filament sliding | ISO | J:73065 | |||||||||
| Biological Process | GO:0030010 | establishment of cell polarity | IMP | J:162326 | |||||||||
| Biological Process | GO:0072673 | lamellipodium morphogenesis | IBA | J:265628 | |||||||||
| Biological Process | GO:0072673 | lamellipodium morphogenesis | IMP | J:162326 | |||||||||
| Biological Process | GO:0048246 | macrophage chemotaxis | IMP | J:162326 | |||||||||
| Biological Process | GO:0002548 | monocyte chemotaxis | IMP | J:162326 | |||||||||
| Biological Process | GO:0043547 | positive regulation of GTPase activity | IDA | J:124018 | |||||||||
| Biological Process | GO:0043547 | positive regulation of GTPase activity | IMP | J:162326 | |||||||||
| Biological Process | GO:0035023 | regulation of Rho protein signal transduction | ISO | J:164563 | |||||||||
| Biological Process | GO:0007266 | Rho protein signal transduction | ISO | J:155856 | |||||||||
| Biological Process | GO:0007266 | Rho protein signal transduction | IMP | J:162326 | |||||||||
| Biological Process | GO:0035385 | Roundabout signaling pathway | ISO | J:164563 | |||||||||
| Biological Process | GO:0007165 | signal transduction | IEA | J:72247 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/23/2024 MGI 6.23 |

|
|
|
||