|
Symbol Name ID |
G6pd2
glucose-6-phosphate dehydrogenase 2 MGI:105977 |
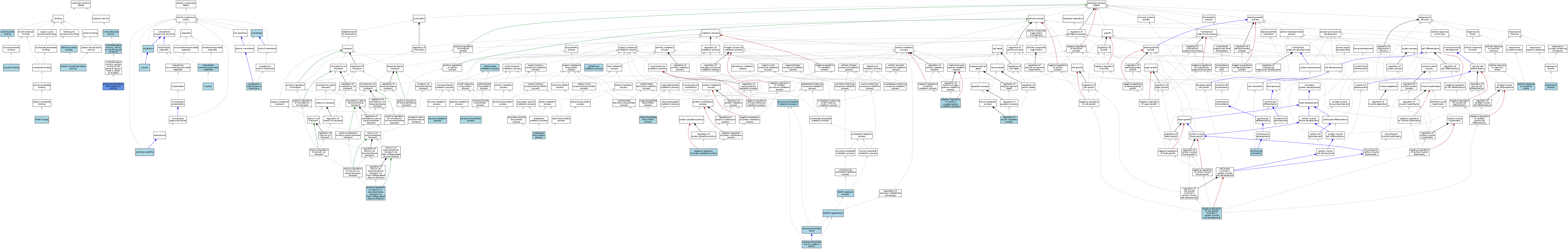
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0030246 | carbohydrate binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0005536 | glucose binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0005536 | glucose binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0004345 | glucose-6-phosphate dehydrogenase activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0004345 | glucose-6-phosphate dehydrogenase activity | ISO | J:155856 | |||||||||
| Molecular Function | GO:0004345 | glucose-6-phosphate dehydrogenase activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0004345 | glucose-6-phosphate dehydrogenase activity | IDA | J:40148 | |||||||||
| Molecular Function | GO:0042802 | identical protein binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0050661 | NADP binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0050661 | NADP binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0016491 | oxidoreductase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0016614 | oxidoreductase activity, acting on CH-OH group of donors | IEA | J:72247 | |||||||||
| Molecular Function | GO:0042803 | protein homodimerization activity | ISO | J:164563 | |||||||||
| Cellular Component | GO:0034451 | centriolar satellite | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0009898 | cytoplasmic side of plasma membrane | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005829 | cytosol | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005829 | cytosol | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005829 | cytosol | IBA | J:265628 | |||||||||
| Cellular Component | GO:0043231 | intracellular membrane-bounded organelle | ISO | J:164563 | |||||||||
| Cellular Component | GO:0043231 | intracellular membrane-bounded organelle | ISO | J:155856 | |||||||||
| Cellular Component | GO:0016020 | membrane | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005634 | nucleus | ISO | J:155856 | |||||||||
| Biological Process | GO:0005975 | carbohydrate metabolic process | IEA | J:60000 | |||||||||
| Biological Process | GO:0034599 | cellular response to oxidative stress | ISO | J:164563 | |||||||||
| Biological Process | GO:0006695 | cholesterol biosynthetic process | ISO | J:164563 | |||||||||
| Biological Process | GO:0043249 | erythrocyte maturation | ISO | J:164563 | |||||||||
| Biological Process | GO:0051156 | glucose 6-phosphate metabolic process | ISO | J:155856 | |||||||||
| Biological Process | GO:0051156 | glucose 6-phosphate metabolic process | ISO | J:164563 | |||||||||
| Biological Process | GO:0006006 | glucose metabolic process | IBA | J:265628 | |||||||||
| Biological Process | GO:0006749 | glutathione metabolic process | ISO | J:164563 | |||||||||
| Biological Process | GO:0006739 | NADP metabolic process | ISO | J:164563 | |||||||||
| Biological Process | GO:0006740 | NADPH regeneration | ISO | J:164563 | |||||||||
| Biological Process | GO:0061052 | negative regulation of cell growth involved in cardiac muscle cell development | ISO | J:155856 | |||||||||
| Biological Process | GO:0010734 | negative regulation of protein glutathionylation | ISO | J:164563 | |||||||||
| Biological Process | GO:2000378 | negative regulation of reactive oxygen species metabolic process | ISO | J:155856 | |||||||||
| Biological Process | GO:0019322 | pentose biosynthetic process | ISO | J:164563 | |||||||||
| Biological Process | GO:0006098 | pentose-phosphate shunt | ISO | J:155856 | |||||||||
| Biological Process | GO:0006098 | pentose-phosphate shunt | ISO | J:164563 | |||||||||
| Biological Process | GO:0009051 | pentose-phosphate shunt, oxidative branch | ISO | J:164563 | |||||||||
| Biological Process | GO:0009051 | pentose-phosphate shunt, oxidative branch | ISO | J:155856 | |||||||||
| Biological Process | GO:0009051 | pentose-phosphate shunt, oxidative branch | IBA | J:265628 | |||||||||
| Biological Process | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel | ISO | J:155856 | |||||||||
| Biological Process | GO:0043523 | regulation of neuron apoptotic process | ISO | J:155856 | |||||||||
| Biological Process | GO:0045471 | response to ethanol | ISO | J:155856 | |||||||||
| Biological Process | GO:0046390 | ribose phosphate biosynthetic process | ISO | J:164563 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/16/2024 MGI 6.23 |

|
|
|
||