|
Symbol Name ID |
Tiam1
T cell lymphoma invasion and metastasis 1 MGI:103306 |
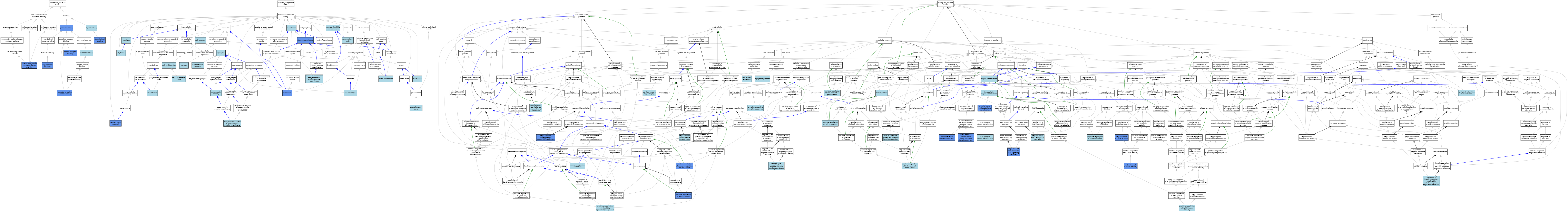
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0046875 | ephrin receptor binding | IPI | J:88877 | |||||||||
| Molecular Function | GO:0046875 | ephrin receptor binding | IPI | J:88877 | |||||||||
| Molecular Function | GO:0005085 | guanyl-nucleotide exchange factor activity | IMP | J:192848 | |||||||||
| Molecular Function | GO:0005085 | guanyl-nucleotide exchange factor activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0005085 | guanyl-nucleotide exchange factor activity | TAS | Reactome:R-NUL-9033341 | |||||||||
| Molecular Function | GO:0005085 | guanyl-nucleotide exchange factor activity | IDA | J:82578 | |||||||||
| Molecular Function | GO:0019900 | kinase binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0008289 | lipid binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0008017 | microtubule binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0008017 | microtubule binding | IDA | J:195272 | |||||||||
| Molecular Function | GO:0005543 | phospholipid binding | IDA | J:82578 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:192848 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:192848 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:156410 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:192848 | |||||||||
| Molecular Function | GO:0030971 | receptor tyrosine kinase binding | IPI | J:180470 | |||||||||
| Molecular Function | GO:0030971 | receptor tyrosine kinase binding | ISO | J:155856 | |||||||||
| Cellular Component | GO:0044295 | axonal growth cone | ISO | J:155856 | |||||||||
| Cellular Component | GO:0030054 | cell junction | IEA | J:60000 | |||||||||
| Cellular Component | GO:0044291 | cell-cell contact zone | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005911 | cell-cell junction | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005829 | cytosol | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005829 | cytosol | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005829 | cytosol | TAS | Reactome:R-NUL-9033304 | |||||||||
| Cellular Component | GO:0005829 | cytosol | TAS | Reactome:R-NUL-9033333 | |||||||||
| Cellular Component | GO:0005829 | cytosol | TAS | Reactome:R-NUL-9033341 | |||||||||
| Cellular Component | GO:0005829 | cytosol | IBA | J:265628 | |||||||||
| Cellular Component | GO:0043197 | dendritic spine | ISO | J:155856 | |||||||||
| Cellular Component | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane | ISO | J:164563 | |||||||||
| Cellular Component | GO:0099147 | extrinsic component of postsynaptic density membrane | ISO | J:155856 | |||||||||
| Cellular Component | GO:0098978 | glutamatergic synapse | ISO | J:155856 | |||||||||
| Cellular Component | GO:0060091 | kinocilium | IDA | J:195272 | |||||||||
| Cellular Component | GO:0044304 | main axon | ISO | J:155856 | |||||||||
| Cellular Component | GO:0016020 | membrane | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005874 | microtubule | ISO | J:155856 | |||||||||
| Cellular Component | GO:0043025 | neuronal cell body | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005634 | nucleus | ISO | J:155856 | |||||||||
| Cellular Component | GO:0000242 | pericentriolar material | IDA | J:195272 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | IDA | J:82578 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | IDA | J:193444 | |||||||||
| Cellular Component | GO:0014069 | postsynaptic density | ISO | J:155856 | |||||||||
| Cellular Component | GO:0032587 | ruffle membrane | ISO | J:155856 | |||||||||
| Cellular Component | GO:0036477 | somatodendritic compartment | ISO | J:155856 | |||||||||
| Cellular Component | GO:0045202 | synapse | ISO | J:155856 | |||||||||
| Cellular Component | GO:0045202 | synapse | IBA | J:265628 | |||||||||
| Biological Process | GO:0090630 | activation of GTPase activity | ISO | J:164563 | |||||||||
| Biological Process | GO:0090630 | activation of GTPase activity | IMP | J:192848 | |||||||||
| Biological Process | GO:0090630 | activation of GTPase activity | ISO | J:155856 | |||||||||
| Biological Process | GO:0090630 | activation of GTPase activity | IBA | J:265628 | |||||||||
| Biological Process | GO:0006915 | apoptotic process | ISO | J:155856 | |||||||||
| Biological Process | GO:0031547 | brain-derived neurotrophic factor receptor signaling pathway | IDA | J:180470 | |||||||||
| Biological Process | GO:0003300 | cardiac muscle hypertrophy | ISO | J:155856 | |||||||||
| Biological Process | GO:0016477 | cell migration | ISO | J:164563 | |||||||||
| Biological Process | GO:0007160 | cell-matrix adhesion | ISO | J:164563 | |||||||||
| Biological Process | GO:0048013 | ephrin receptor signaling pathway | IDA | J:88877 | |||||||||
| Biological Process | GO:0035556 | intracellular signal transduction | IEA | J:72247 | |||||||||
| Biological Process | GO:1990138 | neuron projection extension | ISO | J:155856 | |||||||||
| Biological Process | GO:0098989 | NMDA selective glutamate receptor signaling pathway | ISO | J:155856 | |||||||||
| Biological Process | GO:0050772 | positive regulation of axonogenesis | ISO | J:155856 | |||||||||
| Biological Process | GO:0050772 | positive regulation of axonogenesis | IBA | J:265628 | |||||||||
| Biological Process | GO:0050772 | positive regulation of axonogenesis | IMP | J:88877 | |||||||||
| Biological Process | GO:0030335 | positive regulation of cell migration | ISO | J:164563 | |||||||||
| Biological Process | GO:0030335 | positive regulation of cell migration | ISO | J:155856 | |||||||||
| Biological Process | GO:0008284 | positive regulation of cell population proliferation | ISO | J:164563 | |||||||||
| Biological Process | GO:0061003 | positive regulation of dendritic spine morphogenesis | ISO | J:155856 | |||||||||
| Biological Process | GO:0043507 | positive regulation of JUN kinase activity | ISO | J:155856 | |||||||||
| Biological Process | GO:0010976 | positive regulation of neuron projection development | ISO | J:155856 | |||||||||
| Biological Process | GO:0010976 | positive regulation of neuron projection development | IDA | J:180470 | |||||||||
| Biological Process | GO:0032092 | positive regulation of protein binding | ISO | J:164563 | |||||||||
| Biological Process | GO:1904268 | positive regulation of Schwann cell chemotaxis | ISO | J:155856 | |||||||||
| Biological Process | GO:0072657 | protein localization to membrane | ISO | J:155856 | |||||||||
| Biological Process | GO:0065003 | protein-containing complex assembly | ISO | J:164563 | |||||||||
| Biological Process | GO:0016601 | Rac protein signal transduction | ISO | J:164563 | |||||||||
| Biological Process | GO:1904338 | regulation of dopaminergic neuron differentiation | IMP | J:192848 | |||||||||
| Biological Process | GO:0010717 | regulation of epithelial to mesenchymal transition | ISO | J:164563 | |||||||||
| Biological Process | GO:0070372 | regulation of ERK1 and ERK2 cascade | ISO | J:155856 | |||||||||
| Biological Process | GO:0043087 | regulation of GTPase activity | IDA | J:180470 | |||||||||
| Biological Process | GO:0061178 | regulation of insulin secretion involved in cellular response to glucose stimulus | ISO | J:155856 | |||||||||
| Biological Process | GO:1905274 | regulation of modification of postsynaptic actin cytoskeleton | ISO | J:155856 | |||||||||
| Biological Process | GO:2000050 | regulation of non-canonical Wnt signaling pathway | IGI | J:192848 | |||||||||
| Biological Process | GO:0099175 | regulation of postsynapse organization | ISO | J:155856 | |||||||||
| Biological Process | GO:0007165 | signal transduction | IEA | J:72247 | |||||||||
| Biological Process | GO:0007264 | small GTPase mediated signal transduction | IBA | J:265628 | |||||||||
| Biological Process | GO:0007264 | small GTPase mediated signal transduction | IDA | J:82578 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/23/2024 MGI 6.23 |

|
|
|
||