Comparative GO Graph (mouse, human, rat, zebrafish)
Compare experimental GO annotations for Human-Mouse-Rat-Zebrafish orthologs to Mouse Ubl4a
Graphs display curated GO classifications for mouse, human, rat and zebrafish orthologs annotated from the biomedical literature.
- This class contains
- Human genes UBL4A
- Mouse genes Ubl4a
- Rat genes Ubl4a
- Zfish genes zgc:56596
- Annotations are indicated with nodes colored by organism:
Human,
Mouse,
Rat,
Zfish
- Relations between GO terms are indicated by colored edges "is_a"; "part_of"; "regulates"; "positively_regulates"; "negatively_regulates"
- GO annotations: Human from GOA; Mouse from MGI; Rat from RGD; Zebrafish from ZFIN.
- Only experimental annotations are displayed: EXP, IDA, IEP, IGI, IMP, IPI, HTP, HDA, HMP, HGI, HEP. Evidence codes are listed at the bottom of the page.
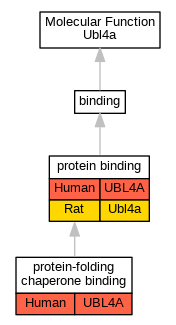
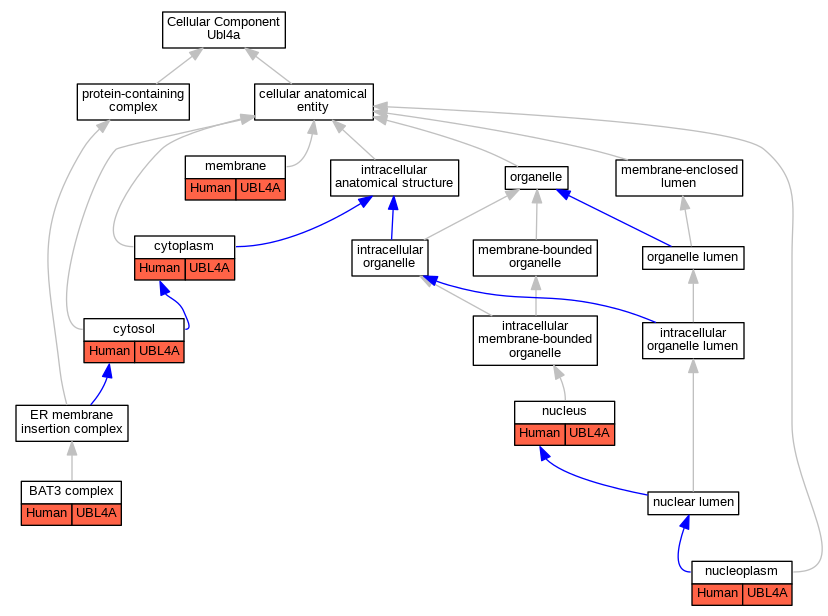
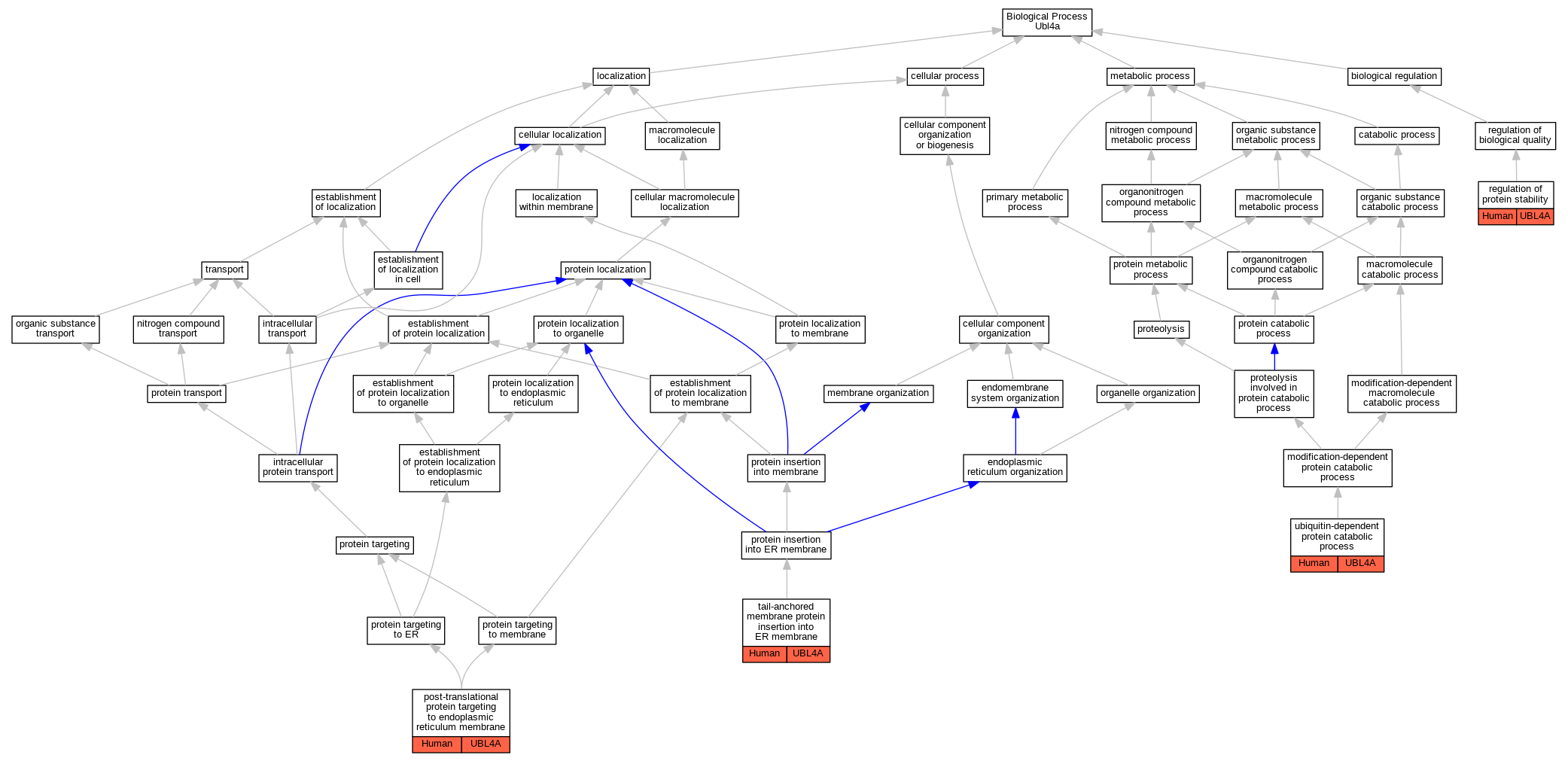 Full set of experimental annotations for Human-Rat-Zebrafish genes considered orthologs of Mouse in the Alliance stringent orthology set Ubl4a
Full set of experimental annotations for Human-Rat-Zebrafish genes considered orthologs of Mouse in the Alliance stringent orthology set Ubl4a
| Category |
ID |
Classification term |
Gene |
Evidence |
Inferred from |
Organism |
Reference |
| Molecular Function | GO:0005515 | protein binding | UBL4A | IPI | UniProtKB:O43765 | Human | PMID:19615732 |
| Molecular Function | GO:0005515 | protein binding | UBL4A | IPI | UniProtKB:P12504 | Human | PMID:22190034 |
| Molecular Function | GO:0005515 | protein binding | UBL4A | IPI | UniProtKB:O43765 | Human | PMID:23246001 |
| Molecular Function | GO:0005515 | protein binding | UBL4A | IPI | UniProtKB:O43765 | Human | PMID:25036637 |
| Molecular Function | GO:0005515 | protein binding | UBL4A | IPI | UniProtKB:P50222 | Human | PMID:25416956 |
| Molecular Function | GO:0005515 | protein binding | UBL4A | IPI | UniProtKB:Q96GN5 | Human | PMID:25416956 |
| Molecular Function | GO:0005515 | protein binding | UBL4A | IPI | UniProtKB:Q9BUZ4 | Human | PMID:25416956 |
| Molecular Function | GO:0005515 | protein binding | UBL4A | IPI | UniProtKB:P46379 | Human | PMID:25713138 |
| Molecular Function | GO:0005515 | protein binding | UBL4A | IPI | UniProtKB:Q96EQ0 | Human | PMID:26496610 |
| Molecular Function | GO:0005515 | protein binding | UBL4A | IPI | UniProtKB:O15055 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | UBL4A | IPI | UniProtKB:O43765 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | UBL4A | IPI | UniProtKB:P25815 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | UBL4A | IPI | UniProtKB:Q96DN0 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | UBL4A | IPI | UniProtKB:Q96EQ0 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | UBL4A | IPI | UniProtKB:Q9BUZ4 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | UBL4A | IPI | UniProtKB:Q9BYV2 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | UBL4A | IPI | UniProtKB:P22607 | Human | PMID:32814053 |
| Molecular Function | GO:0005515 | protein binding | UBL4A | IPI | UniProtKB:P28329-3 | Human | PMID:32814053 |
| Molecular Function | GO:0005515 | protein binding | Ubl4a | IPI | UniProtKB:A0A0G2K916,UniProtKB:B5DFN4,UniProtKB:D3Z941,UniProtKB:D4AA63,UniProtKB:F1M853,UniProtKB:P11250,UniProtKB:P51593,UniProtKB:P61107,UniProtKB:P62718,UniProtKB:P97612,UniProtKB:Q09167,UniProtKB:Q498M4,UniProtKB:Q5PPJ6,UniProtKB:Q5PPK9,UniProtKB:Q63569,UniProtKB:Q6AYJ7 | Rat | PMID:27191843|RGD:11344934 |
| Molecular Function | GO:0051087 | protein-folding chaperone binding | UBL4A | IPI | UniProtKB:P46379 | Human | PMID:25535373 |
| Cellular Component | GO:0071818 | BAT3 complex | UBL4A | IPI | | Human | PMID:25535373 |
| Cellular Component | GO:0071818 | BAT3 complex | UBL4A | IDA | | Human | PMID:20676083 |
| Cellular Component | GO:0071818 | BAT3 complex | UBL4A | IDA | | Human | PMID:21636303 |
| Cellular Component | GO:0071818 | BAT3 complex | UBL4A | IDA | | Human | PMID:25535373 |
| Cellular Component | GO:0005737 | cytoplasm | UBL4A | IDA | | Human | PMID:21636303 |
| Cellular Component | GO:0005829 | cytosol | UBL4A | IDA | | Human | GO_REF:0000052 |
| Cellular Component | GO:0005829 | cytosol | UBL4A | IDA | | Human | PMID:20676083 |
| Cellular Component | GO:0005829 | cytosol | UBL4A | IDA | | Human | PMID:23246001 |
| Cellular Component | GO:0016020 | membrane | UBL4A | IDA | | Human | PMID:23246001 |
| Cellular Component | GO:0005654 | nucleoplasm | UBL4A | IDA | | Human | GO_REF:0000052 |
| Cellular Component | GO:0005634 | nucleus | UBL4A | IDA | | Human | PMID:21636303 |
| Biological Process | GO:0006620 | post-translational protein targeting to endoplasmic reticulum membrane | UBL4A | IDA | | Human | PMID:25535373 |
| Biological Process | GO:0031647 | regulation of protein stability | UBL4A | IDA | | Human | PMID:21636303 |
| Biological Process | GO:0071816 | tail-anchored membrane protein insertion into ER membrane | UBL4A | IMP | | Human | PMID:20676083 |
| Biological Process | GO:0071816 | tail-anchored membrane protein insertion into ER membrane | UBL4A | IDA | | Human | PMID:25535373 |
| Biological Process | GO:0006511 | ubiquitin-dependent protein catabolic process | UBL4A | IDA | | Human | PMID:20676083 |
Gene Ontology Evidence Code Abbreviations:
| EXP Inferred from experiment |
| IDA Inferred from direct assay |
| IEP Inferred from expression pattern |
| IGI Inferred from genetic interaction |
| IMP Inferred from mutant phenotype |
| IPI Inferred from physical interaction |
| HTP Inferred from High Throughput Experiment |
| HDA Inferred from High Throughput Direct Assay |
| HMP Inferred from High Throughput Mutant Phenotype |
| HGI Inferred from High Throughput Genetic Interaction |
| HEP Inferred from High Throughput Expression Pattern |
 Analysis Tools
Analysis Tools





