Comparative GO Graph (mouse, human, rat, zebrafish)
Compare experimental GO annotations for Human-Mouse-Rat-Zebrafish orthologs to Mouse Clrn1
Graphs display curated GO classifications for mouse, human, rat and zebrafish orthologs annotated from the biomedical literature.
- This class contains
- Human genes CLRN1
- Mouse genes Clrn1
- Rat genes Clrn1
- Zfish genes clrn1
- Annotations are indicated with nodes colored by organism:
Human,
Mouse,
Rat,
Zfish
- Relations between GO terms are indicated by colored edges "is_a"; "part_of"; "regulates"; "positively_regulates"; "negatively_regulates"
- GO annotations: Human from GOA; Mouse from MGI; Rat from RGD; Zebrafish from ZFIN.
- Only experimental annotations are displayed: EXP, IDA, IEP, IGI, IMP, IPI, HTP, HDA, HMP, HGI, HEP. Evidence codes are listed at the bottom of the page.
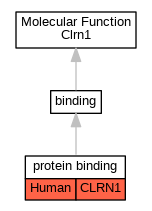
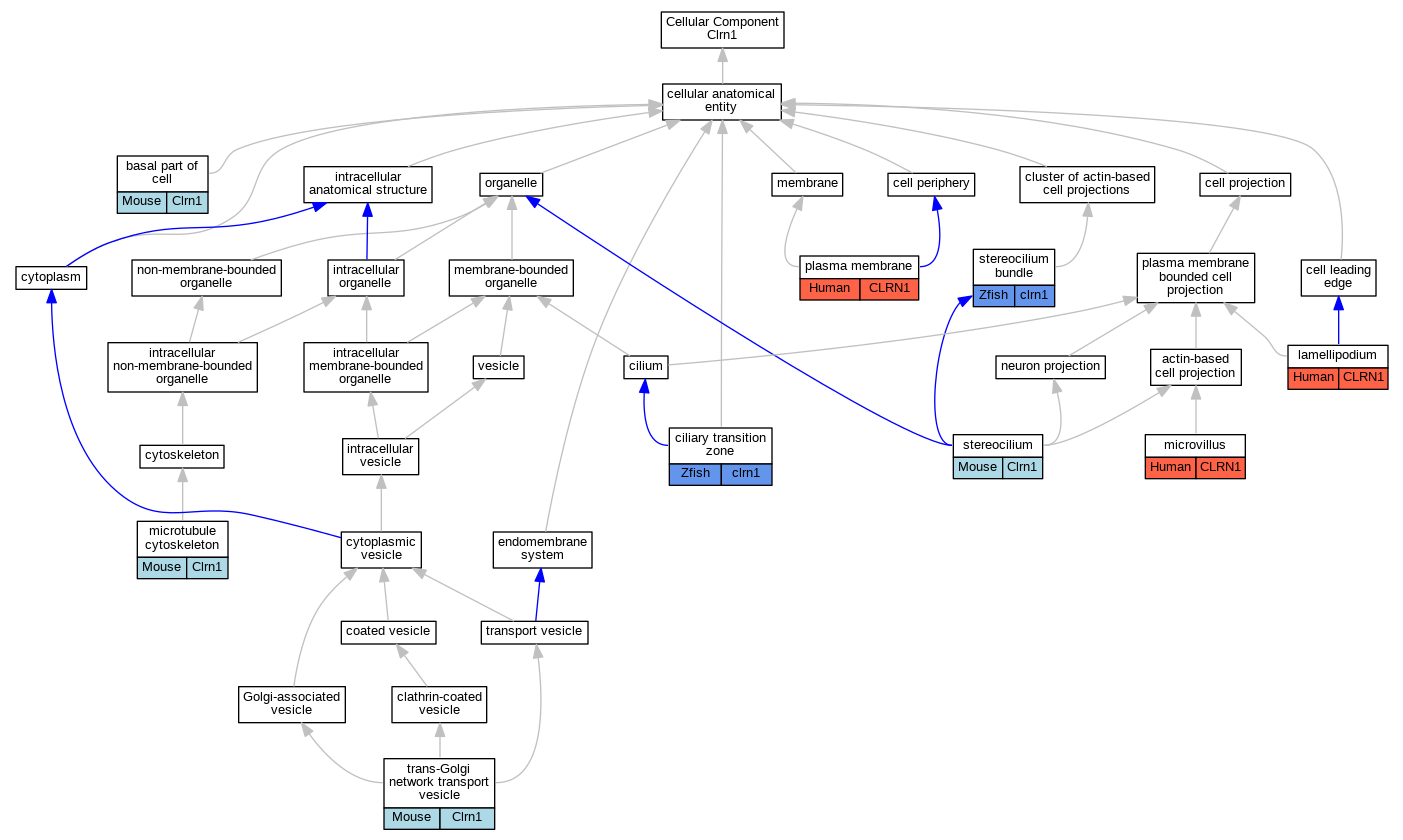
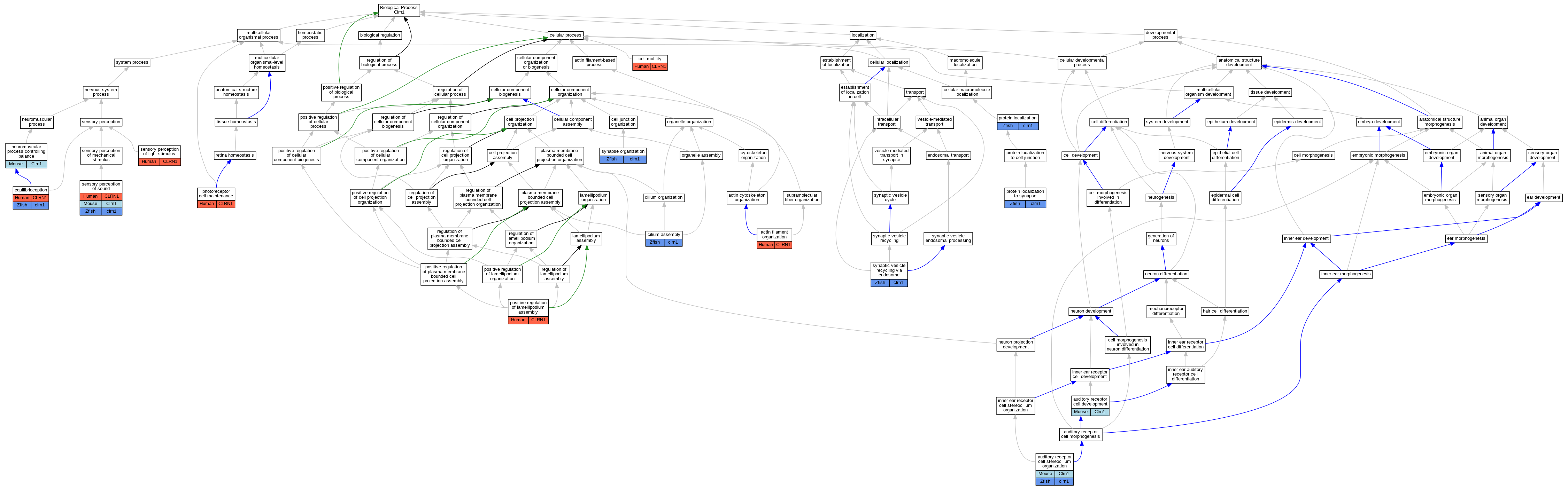 Full set of experimental annotations for Human-Rat-Zebrafish genes considered orthologs of Mouse in the Alliance stringent orthology set Clrn1
Full set of experimental annotations for Human-Rat-Zebrafish genes considered orthologs of Mouse in the Alliance stringent orthology set Clrn1
| Category |
ID |
Classification term |
Gene |
Evidence |
Inferred from |
Organism |
Reference |
| Molecular Function | GO:0005515 | protein binding | CLRN1 | IPI | UniProtKB:O14523 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | CLRN1 | IPI | UniProtKB:O14880 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | CLRN1 | IPI | UniProtKB:O75396 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | CLRN1 | IPI | UniProtKB:P11215 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | CLRN1 | IPI | UniProtKB:P56557 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | CLRN1 | IPI | UniProtKB:Q4LDR2 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | CLRN1 | IPI | UniProtKB:Q53HI1 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | CLRN1 | IPI | UniProtKB:Q6N075 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | CLRN1 | IPI | UniProtKB:Q6PL45-2 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | CLRN1 | IPI | UniProtKB:Q6ZT21 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | CLRN1 | IPI | UniProtKB:Q8IXM6 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | CLRN1 | IPI | UniProtKB:Q8N130 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | CLRN1 | IPI | UniProtKB:Q8N5W8 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | CLRN1 | IPI | UniProtKB:Q8N661 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | CLRN1 | IPI | UniProtKB:Q8N682 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | CLRN1 | IPI | UniProtKB:Q8N6F1-2 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | CLRN1 | IPI | UniProtKB:Q8NHW4 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | CLRN1 | IPI | UniProtKB:Q8WVQ1 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | CLRN1 | IPI | UniProtKB:Q8WW34-2 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | CLRN1 | IPI | UniProtKB:Q96MV8 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | CLRN1 | IPI | UniProtKB:Q9BVK2 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | CLRN1 | IPI | UniProtKB:Q9BVK8 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | CLRN1 | IPI | UniProtKB:Q9H0R3 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | CLRN1 | IPI | UniProtKB:Q9NRS4 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | CLRN1 | IPI | UniProtKB:Q9NV12 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | CLRN1 | IPI | UniProtKB:Q9Y231 | Human | PMID:32296183 |
| Cellular Component | GO:0045178 | basal part of cell | Clrn1 | IDA | | Mouse | PMID:19539019 |
| Cellular Component | GO:0035869 | ciliary transition zone | clrn1 | IDA | | Zfish | PMID:25365995 |
| Cellular Component | GO:0030027 | lamellipodium | CLRN1 | IDA | | Human | PMID:19423712 |
| Cellular Component | GO:0015630 | microtubule cytoskeleton | Clrn1 | IDA | | Mouse | PMID:19539019 |
| Cellular Component | GO:0005902 | microvillus | CLRN1 | IDA | | Human | PMID:19423712 |
| Cellular Component | GO:0005886 | plasma membrane | CLRN1 | IDA | | Human | PMID:19423712 |
| Cellular Component | GO:0032420 | stereocilium | Clrn1 | IDA | | Mouse | PMID:19539019 |
| Cellular Component | GO:0032421 | stereocilium bundle | clrn1 | IDA | | Zfish | PMID:26180195 |
| Cellular Component | GO:0030140 | trans-Golgi network transport vesicle | Clrn1 | IDA | | Mouse | PMID:19539019 |
| Biological Process | GO:0007015 | actin filament organization | CLRN1 | IDA | | Human | PMID:19423712 |
| Biological Process | GO:0060117 | auditory receptor cell development | Clrn1 | IMP | MGI:MGI:3850171 | Mouse | PMID:19414487 |
| Biological Process | GO:0060088 | auditory receptor cell stereocilium organization | Clrn1 | IMP | MGI:MGI:3850171 | Mouse | PMID:19414487 |
| Biological Process | GO:0060088 | auditory receptor cell stereocilium organization | Clrn1 | IMP | MGI:MGI:3850171 | Mouse | PMID:19423712 |
| Biological Process | GO:0060088 | auditory receptor cell stereocilium organization | clrn1 | IMP | ZFIN:ZDB-GENO-161111-3 | Zfish | PMID:31097578 |
| Biological Process | GO:0048870 | cell motility | CLRN1 | IDA | | Human | PMID:19423712 |
| Biological Process | GO:0060271 | cilium assembly | clrn1 | IMP | ZFIN:ZDB-MRPHLNO-150211-3,ZFIN:ZDB-MRPHLNO-150211-4,ZFIN:ZDB-MRPHLNO-150211-5 | Zfish | PMID:25365995 |
| Biological Process | GO:0050957 | equilibrioception | CLRN1 | IMP | | Human | PMID:15521980 |
| Biological Process | GO:0050957 | equilibrioception | clrn1 | IMP | ZFIN:ZDB-GENO-161111-1 | Zfish | PMID:26180195 |
| Biological Process | GO:0050885 | neuromuscular process controlling balance | Clrn1 | IMP | MGI:MGI:3850171 | Mouse | PMID:19414487 |
| Biological Process | GO:0045494 | photoreceptor cell maintenance | CLRN1 | IMP | | Human | PMID:15521980 |
| Biological Process | GO:0010592 | positive regulation of lamellipodium assembly | CLRN1 | IDA | | Human | PMID:19423712 |
| Biological Process | GO:0008104 | protein localization | clrn1 | IGI | ZFIN:ZDB-MRPHLNO-150211-6 | Zfish | PMID:25365995 |
| Biological Process | GO:0008104 | protein localization | clrn1 | IMP | ZFIN:ZDB-MRPHLNO-150211-3,ZFIN:ZDB-MRPHLNO-150211-4,ZFIN:ZDB-MRPHLNO-150211-5 | Zfish | PMID:25365995 |
| Biological Process | GO:0035418 | protein localization to synapse | clrn1 | IMP | ZFIN:ZDB-MRPHLNO-150211-3,ZFIN:ZDB-MRPHLNO-150211-4,ZFIN:ZDB-MRPHLNO-150211-5 | Zfish | PMID:25365995 |
| Biological Process | GO:0050953 | sensory perception of light stimulus | CLRN1 | IMP | | Human | PMID:15521980 |
| Biological Process | GO:0007605 | sensory perception of sound | CLRN1 | IMP | | Human | PMID:15650299 |
| Biological Process | GO:0007605 | sensory perception of sound | Clrn1 | IMP | MGI:MGI:3850171 | Mouse | PMID:19414487 |
| Biological Process | GO:0007605 | sensory perception of sound | clrn1 | IMP | ZFIN:ZDB-GENO-161111-1 | Zfish | PMID:26180195 |
| Biological Process | GO:0050808 | synapse organization | clrn1 | IMP | ZFIN:ZDB-MRPHLNO-150211-3,ZFIN:ZDB-MRPHLNO-150211-4,ZFIN:ZDB-MRPHLNO-150211-5 | Zfish | PMID:25365995 |
| Biological Process | GO:0036466 | synaptic vesicle recycling via endosome | clrn1 | IMP | ZFIN:ZDB-MRPHLNO-150211-3,ZFIN:ZDB-MRPHLNO-150211-4,ZFIN:ZDB-MRPHLNO-150211-5 | Zfish | PMID:25365995 |
Gene Ontology Evidence Code Abbreviations:
| EXP Inferred from experiment |
| IDA Inferred from direct assay |
| IEP Inferred from expression pattern |
| IGI Inferred from genetic interaction |
| IMP Inferred from mutant phenotype |
| IPI Inferred from physical interaction |
| HTP Inferred from High Throughput Experiment |
| HDA Inferred from High Throughput Direct Assay |
| HMP Inferred from High Throughput Mutant Phenotype |
| HGI Inferred from High Throughput Genetic Interaction |
| HEP Inferred from High Throughput Expression Pattern |
 Analysis Tools
Analysis Tools





