Diagrams of general structure of IKMC primary alleles and derivative alleles
The IKMC (International Knockout Mouse Consortium) created a large resource of mutant ES cell lines using large-scale production pipelines. Alleles of certain types, made with defined vectors, were generated. These diagrams illustrate the most common of these constructs.
See Nomenclature for mutant alleles generated in ES cell lines by the IKMC for details on nomenclature.
There were 5 primary classes of mutant alleles produced by the IKMC (all in ES cell lines). In some cases, derivative alleles can be generated from the primary allele. In addition, targeted cre knockin alleles are being made in conjunction with IKMC by the EUCOMMtools project.
- Gene Trapped Alleles
- Targeted Reporter-tagged Deletion Alleles
- Targeted Knockout First, Reporter-tagged Insertion with Conditional Potential (conditional-ready)
- Targeted Knockout First, Reporter-tagged Insertion NON-conditional
- Targeted Knockout First Alleles with Artificial Introns
- Targeted cre Knockin Alleles

1. Gene Trapped Alleles.
| 1.1 Gene trapped alleles made by Texas A&M Institute for Genomic Medicine for the IKMC program were of two types, differing in whether they carried a beta-geo or neo cassette. |
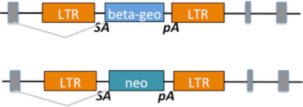 |
Example allele:
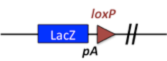
| 1.2 Gene trapped alleles made by EUCOMM include multiple
FRT and loxP sites, making it possible to generate several derivative alleles. |
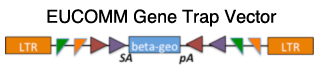 |
Example allele:
| Nmnat2Gt(EUCE0262a08)Hmgu |
nicotinamide nucleotide adenylyltransferase 2; gene trap EUCE0262a08, Helmholtz Zentrum Muenchen GmbH |
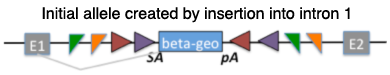 |
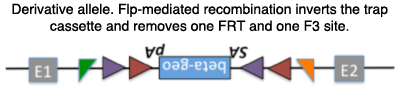
Derivative alleles from the EUCOMM gene traps can be made, taking advantage of the FRT and loxP sites. One such derivative, using flp-mediated recombination, inverts the trap cassette allowing for conditional targeting of the endogenous gene using cre.
Example derivative allele:
1.3 Gene trapped alleles made by NorCOMM (NCOM) used a variety of vectors, one of which was UPA, shown below.
Example allele:
| Fgfr2Gt(PST17605)Mfgc |
fibroblast growth factor receptor 2; gene trap PST17605, Mammalian Functional Genomics Centre |
 |
Various derivative alleles can be made, depending on the inclusion of loxP and FRT sites.
Top
2. Targeted Reporter-tagged Deletion Alleles.
| 2.1 Reporter-tagged deletion alleles were developed by Regeneron, Inc. using Velocigene technology. In most cases, these alleles delete the entire coding region of the gene, replacing it with a LacZ, loxP, neo, loxP containing cassette. |
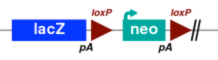 |
Example allele:
A derivative allele can be created from Velocigene alleles by removing the neomycin cassette through cre-excised deletion. The designation for this cre-excised deletion allele changes the tm# portion of the allele symbol to tm#.1
Example derivative allele:
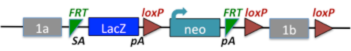
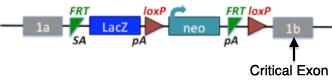
2.2 Reporter-tagged deletion alleles were developed by the Chori/Sanger/Davis collaboration. These alleles delete a critical exon, replacing it with a FRT, LacZ, loxP, neo, FRT, loxP containing cassette.
Example allele:
| Mdktm1(KOMP)Mbp |
midkine; targeted mutation 1, Mouse Biology Program, UCDavis |
[The figure below illustrates the deletion of exon 2 with replacement targeting cassette.]
Example derivative alleles:
[A] Excising the tm1 allele
with [A] flp creates a tm1.2 allele,
where the LacZ and neo are removed. |
[B] Excising the tm1 allele
with [B] cre creates a tm1.1 allele,
where the neo is removed.
|
| | 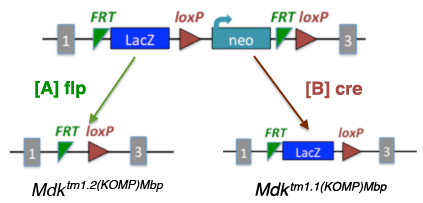 |
2.3 Reporter-tagged deletion alleles developed by the NorCOMM (NCOM) project from the Centre for Modeling Human Disease and the Mammalian Functional Genomics Centre. Two slightly different targeting cassettes were used, each produced LacZ-tagged, conditional-ready alleles that deleted a critical exon. The critical exon can be reintroduced using the attp docking site.
Example alleles:
| Itpr3tm1(NCOM)Mfgc |
inositol 1,4,5-triphosphate receptor 3; targeted mutation 1, Mammalian Functional Genomics Centre |
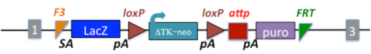 |
| Otud7btm1(NCOM)Cmhd |
OTU domain containing 7B; targeted mutation 1, Centre for Modelling Human Disease |
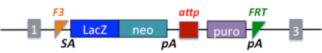 |
Top
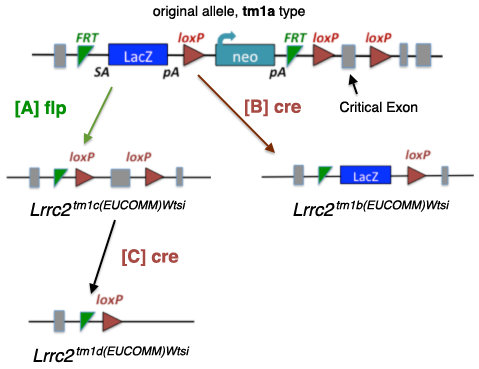
3. Targeted Knockout First, Reporter-tagged Insertion with Conditional Potential (conditional-ready).
These alleles may contain a promoter or be promoterless. Note: Only the promoter-driven versions are diagrammed here. A critical exon is flanked by loxP sites, with FRT, LacZ, loxP, promoter, neo, and FRT elements upstream of the critical exon. Derivative alleles are generated using cre or flp.
Example allele:
| Lrrc2tm1a(EUCOMM)Wtsi |
leucine rich repeat containing 2; targeted mutation 1a,
Wellcome Trust Sanger Institute |
| Example derivative alleles:
[A] Excising the tm1a allele with [A] flp creates a tm1c allele, where the LacZ and neo are removed. The gene is intact, with loxP flanking the critical exon(s), making this an allele suitable for conditional mutagenesis. |
[B] Excising the tm1a type allele with [B] cre creates a tm1b allele, where neo and the
critical exon are removed, leaving a definitive null with a reporter. |
| [C] Excising the tm1c allele with [C] cre removes the critical exon and creates a tm1d allele with only one FRT and one loxP site remaining. |
| |  |
Top
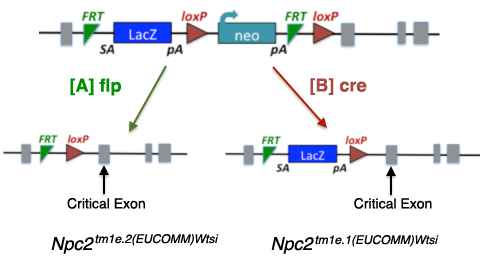
4. Targeted Knockout First, Reporter-tagged Insertion NON-conditional.
These alleles are the same as those described above for the tm1a type allele, except that they have lost the loxP site downstream of the critical exon (CE), so they are no longer conditional. Derivative alleles are generated using cre or flp.
Example allele:
| Plvaptm1e(EUCOMM)Hmgu |
plasmalemma vesicle associated protein; targeted mutation 1e, Helmholtz Zentrum Muenchen GmbH |
| Example derivative alleles:
[A] Excising the tm1e allele with [A] flp creates a tm1e.2 allele type, where the LacZ and neo are removed. |
| [B] Excising the tm1e type allele with [B] cre creates a tm1e.1 allele, where neo is removed.
|
| |  |
Top
5. Targeted Knockout First Alleles with Artificial Introns.
As some genes are very small or contain only a single exon, vector designs to make conditional-ready alleles fail (requires a critical exon). Conditional knockout first alleles for such genes can be made by introducing an artificial intron with the targeting cassette imbedded in the intron. The 'second' exon (1b in the diagram) is then used as the critical exon.
Alleles of type knockout first, reporter-tagged insertion NON-conditional will also arise in cases where the loxP site downstream of the critical exon has been lost (tm1e alleles)
Example alleles:
Derivatives of these alleles can be made, with parallel structures as above, see
- Targeted Knockout First, Reporter-tagged Insertion with Conditional Potential (conditional-ready)
- Targeted Knockout First, Reporter-tagged Insertion NON-conditional
Top
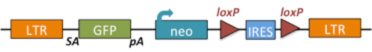
6. Targeted cre Knockin Alleles.
The EUCOMMtools project creates targeted, cre knockin alleles that expand the available worldwide cre resources. The selection cassette is flanked with Rox sites and can be removed using dre recombinase.
| Example allele: |
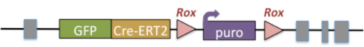 |
| Hprttm1(Cbx1-cre/ERT2)Hmgu |
A hypothetical targeted knockin at the Hprt locus. The knocked in
portion contains promoter sequences from Cbx1 driving cre/ERT2 |
References:
TIGM gene traps: Hansen GM, et al. 2008. Large-scale gene trapping in C57BL/6N mouse embryonic stem cells. Genome Res 18: 1670-1679. PubMed ID: 18799693
EUCOMM gene traps: Schnutgen F, et al. 2005. Genomewide production of multipurpose alleles for the functional analysis of the mouse genome. PNAS 102: 7221-7226. PubMed ID: 15870191
Velocigene Reporter-Tagged Deletion Alleles: Valenzuela DM, et al. 2003. High-throughput engineering of the mouse genome coupled with high-resolution expression analysis. Nat Biotechnol 21: 652-659. PubMed ID: 12730667
Knockout first, reporter-tagged insertion with conditional potential (conditional-ready): Skarnes WC, et al 2011. A conditional knockout resource for the genome-wide study of mouse gene function. Nature 474: 337-432. PubMed ID: 21677750
Overall Review: Bradley A, et al. 2012. The mammalian gene function resource: the International Knockout Mouse Consortium. Mamm Genome 9-10: 580-586. PubMed ID: 22968824
Top
 Analysis Tools
Analysis Tools