|
Symbol Name ID |
Mc1r
melanocortin 1 receptor MGI:99456 |
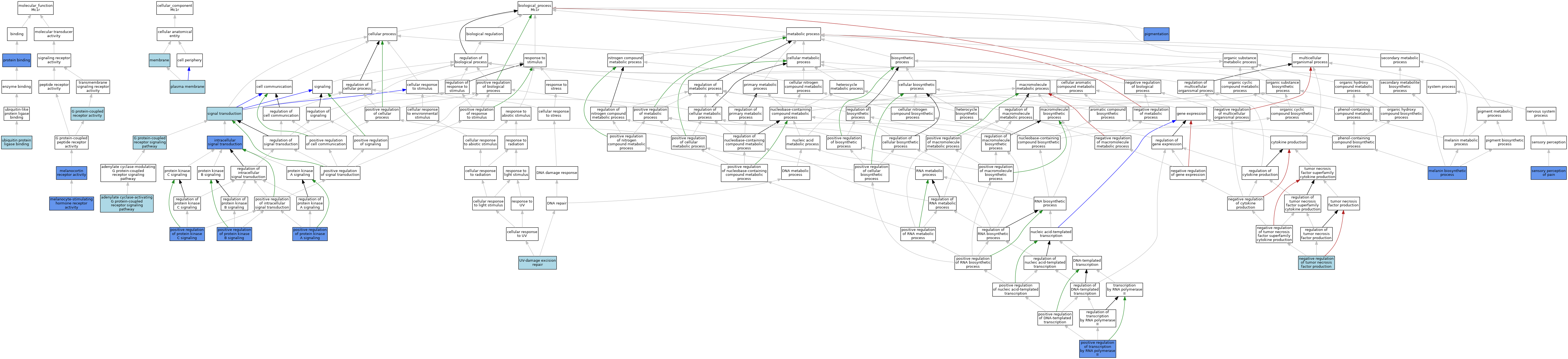
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0004930 | G protein-coupled receptor activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0004930 | G protein-coupled receptor activity | IEA | J:72247 | |||||||||
| Molecular Function | GO:0004977 | melanocortin receptor activity | IDA | J:155191 | |||||||||
| Molecular Function | GO:0004980 | melanocyte-stimulating hormone receptor activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0004980 | melanocyte-stimulating hormone receptor activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0004980 | melanocyte-stimulating hormone receptor activity | IDA | J:45497 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:45497 | |||||||||
| Molecular Function | GO:0031625 | ubiquitin protein ligase binding | ISO | J:164563 | |||||||||
| Cellular Component | GO:0016020 | membrane | IEA | J:72247 | |||||||||
| Cellular Component | GO:0016020 | membrane | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | ISO | J:164563 | |||||||||
| Biological Process | GO:0007189 | adenylate cyclase-activating G protein-coupled receptor signaling pathway | ISO | J:164563 | |||||||||
| Biological Process | GO:0007186 | G protein-coupled receptor signaling pathway | IEA | J:72247 | |||||||||
| Biological Process | GO:0007186 | G protein-coupled receptor signaling pathway | IEA | J:60000 | |||||||||
| Biological Process | GO:0035556 | intracellular signal transduction | IDA | J:155191 | |||||||||
| Biological Process | GO:0042438 | melanin biosynthetic process | IMP | J:28766 | |||||||||
| Biological Process | GO:0032720 | negative regulation of tumor necrosis factor production | ISO | J:164563 | |||||||||
| Biological Process | GO:0043473 | pigmentation | IMP | MGI:MGI:87265 | |||||||||
| Biological Process | GO:0010739 | positive regulation of protein kinase A signaling | IDA | J:155191 | |||||||||
| Biological Process | GO:0051897 | positive regulation of protein kinase B signaling | IDA | J:155191 | |||||||||
| Biological Process | GO:0090037 | positive regulation of protein kinase C signaling | IDA | J:155191 | |||||||||
| Biological Process | GO:0045944 | positive regulation of transcription by RNA polymerase II | IDA | J:155191 | |||||||||
| Biological Process | GO:0019233 | sensory perception of pain | IMP | J:83404 | |||||||||
| Biological Process | GO:0007165 | signal transduction | IEA | J:60000 | |||||||||
| Biological Process | GO:0070914 | UV-damage excision repair | ISO | J:164563 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/16/2024 MGI 6.23 |

|
|
|
||