|
Symbol Name ID |
Vcl
vinculin MGI:98927 |
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
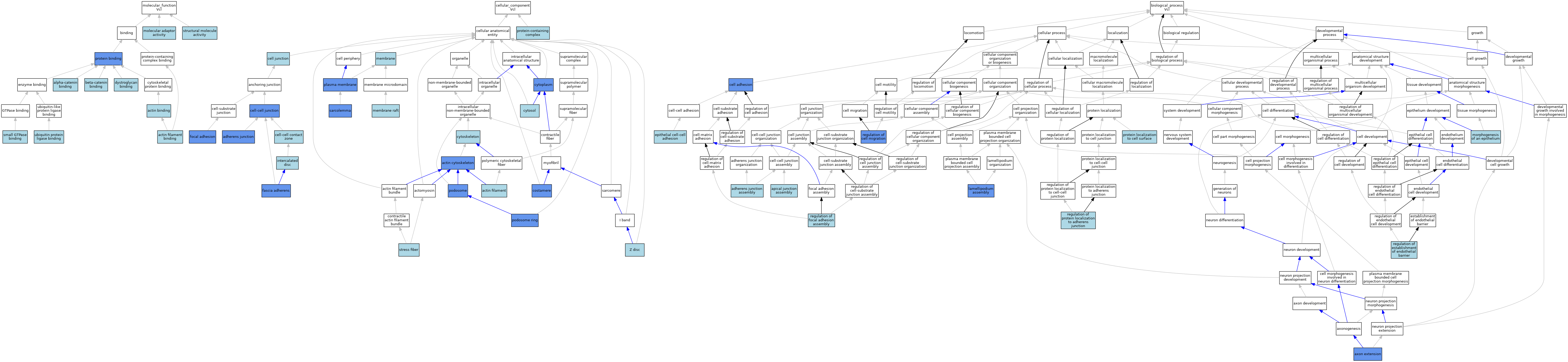
| Molecular Function | GO:0003779 | actin binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0003779 | actin binding | IEA | J:72247 | |||||||||
| Molecular Function | GO:0051015 | actin filament binding | IEA | J:72247 | |||||||||
| Molecular Function | GO:0045294 | alpha-catenin binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0045294 | alpha-catenin binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:0008013 | beta-catenin binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:0002162 | dystroglycan binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0060090 | molecular adaptor activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:200373 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:116379 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:212352 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:200373 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:114108 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:201288 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:193640 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:227201 | |||||||||
| Molecular Function | GO:0031267 | small GTPase binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0005198 | structural molecule activity | IEA | J:72247 | |||||||||
| Molecular Function | GO:0031625 | ubiquitin protein ligase binding | ISO | J:164563 | |||||||||
| Cellular Component | GO:0015629 | actin cytoskeleton | IDA | J:166692 | |||||||||
| Cellular Component | GO:0005884 | actin filament | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005912 | adherens junction | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005912 | adherens junction | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005912 | adherens junction | IDA | J:78855 | |||||||||
| Cellular Component | GO:0030054 | cell junction | IEA | J:60000 | |||||||||
| Cellular Component | GO:0044291 | cell-cell contact zone | ISO | J:164563 | |||||||||
| Cellular Component | GO:0044291 | cell-cell contact zone | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005911 | cell-cell junction | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005911 | cell-cell junction | IDA | J:87902 | |||||||||
| Cellular Component | GO:0005911 | cell-cell junction | IDA | J:191757 | |||||||||
| Cellular Component | GO:0043034 | costamere | ISO | J:73065 | |||||||||
| Cellular Component | GO:0043034 | costamere | IDA | J:91191 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IDA | J:213085 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005856 | cytoskeleton | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005829 | cytosol | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005916 | fascia adherens | IDA | J:112534 | |||||||||
| Cellular Component | GO:0005916 | fascia adherens | IDA | J:73178 | |||||||||
| Cellular Component | GO:0005925 | focal adhesion | IDA | J:213085 | |||||||||
| Cellular Component | GO:0005925 | focal adhesion | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005925 | focal adhesion | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005925 | focal adhesion | IDA | J:114718 | |||||||||
| Cellular Component | GO:0005925 | focal adhesion | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005925 | focal adhesion | IDA | J:227201 | |||||||||
| Cellular Component | GO:0005925 | focal adhesion | IDA | J:217307 | |||||||||
| Cellular Component | GO:0005925 | focal adhesion | ISO | J:254431 | |||||||||
| Cellular Component | GO:0014704 | intercalated disc | ISO | J:155856 | |||||||||
| Cellular Component | GO:0016020 | membrane | IEA | J:60000 | |||||||||
| Cellular Component | GO:0045121 | membrane raft | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | IDA | J:112534 | |||||||||
| Cellular Component | GO:0002102 | podosome | IDA | J:129034 | |||||||||
| Cellular Component | GO:0061826 | podosome ring | IDA | J:246351 | |||||||||
| Cellular Component | GO:0032991 | protein-containing complex | ISO | J:164563 | |||||||||
| Cellular Component | GO:0042383 | sarcolemma | IDA | J:243471 | |||||||||
| Cellular Component | GO:0042383 | sarcolemma | ISO | J:155856 | |||||||||
| Cellular Component | GO:0001725 | stress fiber | ISO | J:155856 | |||||||||
| Cellular Component | GO:0030018 | Z disc | ISO | J:155856 | |||||||||
| Biological Process | GO:0034333 | adherens junction assembly | ISO | J:164563 | |||||||||
| Biological Process | GO:0043297 | apical junction assembly | ISO | J:164563 | |||||||||
| Biological Process | GO:0048675 | axon extension | IGI | J:209602 | |||||||||
| Biological Process | GO:0048675 | axon extension | IGI | J:209602 | |||||||||
| Biological Process | GO:0007155 | cell adhesion | IBA | J:265628 | |||||||||
| Biological Process | GO:0007155 | cell adhesion | IDA | J:80756 | |||||||||
| Biological Process | GO:0090136 | epithelial cell-cell adhesion | ISO | J:164563 | |||||||||
| Biological Process | GO:0030032 | lamellipodium assembly | IDA | J:80756 | |||||||||
| Biological Process | GO:0002009 | morphogenesis of an epithelium | ISO | J:164563 | |||||||||
| Biological Process | GO:0034394 | protein localization to cell surface | ISO | J:164563 | |||||||||
| Biological Process | GO:0030334 | regulation of cell migration | IDA | J:80756 | |||||||||
| Biological Process | GO:1903140 | regulation of establishment of endothelial barrier | ISO | J:164563 | |||||||||
| Biological Process | GO:0051893 | regulation of focal adhesion assembly | ISO | J:164563 | |||||||||
| Biological Process | GO:1904702 | regulation of protein localization to adherens junction | ISO | J:164563 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/30/2024 MGI 6.23 |

|
|
|
||