|
Symbol Name ID |
Ucp1
uncoupling protein 1 (mitochondrial, proton carrier) MGI:98894 |
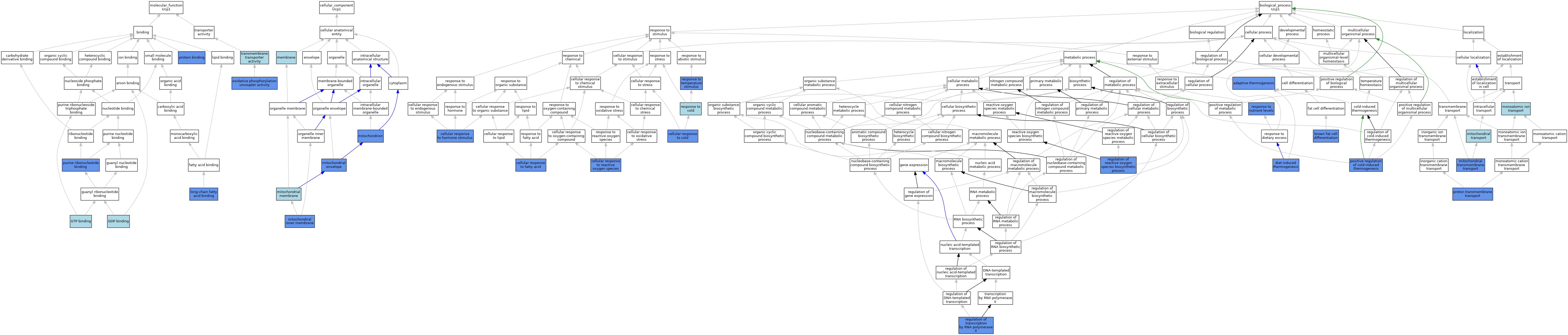
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0019003 | GDP binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0005525 | GTP binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0036041 | long-chain fatty acid binding | IDA | J:189069 | |||||||||
| Molecular Function | GO:0017077 | oxidative phosphorylation uncoupler activity | IMP | J:165368 | |||||||||
| Molecular Function | GO:0017077 | oxidative phosphorylation uncoupler activity | ISO | J:155856 | |||||||||
| Molecular Function | GO:0017077 | oxidative phosphorylation uncoupler activity | IDA | J:189069 | |||||||||
| Molecular Function | GO:0017077 | oxidative phosphorylation uncoupler activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0017077 | oxidative phosphorylation uncoupler activity | IDA | J:85229 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:85229 | |||||||||
| Molecular Function | GO:0032555 | purine ribonucleotide binding | IDA | J:189069 | |||||||||
| Molecular Function | GO:0022857 | transmembrane transporter activity | ISO | J:155856 | |||||||||
| Molecular Function | GO:0022857 | transmembrane transporter activity | ISO | J:164563 | |||||||||
| Cellular Component | GO:0016020 | membrane | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005740 | mitochondrial envelope | IDA | J:85229 | |||||||||
| Cellular Component | GO:0005743 | mitochondrial inner membrane | IDA | J:189069 | |||||||||
| Cellular Component | GO:0031966 | mitochondrial membrane | IEA | J:72247 | |||||||||
| Cellular Component | GO:0005739 | mitochondrion | IDA | J:236213 | |||||||||
| Cellular Component | GO:0005739 | mitochondrion | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005739 | mitochondrion | HDA | J:151002 | |||||||||
| Cellular Component | GO:0005739 | mitochondrion | IDA | J:129009 | |||||||||
| Cellular Component | GO:0005739 | mitochondrion | HDA | J:86816 | |||||||||
| Cellular Component | GO:0005739 | mitochondrion | HDA | J:86816 | |||||||||
| Cellular Component | GO:0005739 | mitochondrion | IDA | J:76945 | |||||||||
| Cellular Component | GO:0005739 | mitochondrion | IDA | J:233904 | |||||||||
| Cellular Component | GO:0005739 | mitochondrion | HDA | J:86816 | |||||||||
| Biological Process | GO:1990845 | adaptive thermogenesis | IMP | J:40053 | |||||||||
| Biological Process | GO:1990845 | adaptive thermogenesis | IMP | J:235306 | |||||||||
| Biological Process | GO:1990845 | adaptive thermogenesis | IBA | J:265628 | |||||||||
| Biological Process | GO:0050873 | brown fat cell differentiation | IEP | J:83859 | |||||||||
| Biological Process | GO:0050873 | brown fat cell differentiation | IDA | J:245403 | |||||||||
| Biological Process | GO:0070417 | cellular response to cold | IDA | J:245403 | |||||||||
| Biological Process | GO:0071398 | cellular response to fatty acid | IDA | J:189069 | |||||||||
| Biological Process | GO:0071398 | cellular response to fatty acid | ISO | J:155856 | |||||||||
| Biological Process | GO:0032870 | cellular response to hormone stimulus | IMP | J:235306 | |||||||||
| Biological Process | GO:0034614 | cellular response to reactive oxygen species | IMP | J:235306 | |||||||||
| Biological Process | GO:0002024 | diet induced thermogenesis | IMP | J:146646 | |||||||||
| Biological Process | GO:1990542 | mitochondrial transmembrane transport | IDA | J:189069 | |||||||||
| Biological Process | GO:1990542 | mitochondrial transmembrane transport | IMP | J:165368 | |||||||||
| Biological Process | GO:1990542 | mitochondrial transmembrane transport | ISO | J:155856 | |||||||||
| Biological Process | GO:1990542 | mitochondrial transmembrane transport | IBA | J:265628 | |||||||||
| Biological Process | GO:0006839 | mitochondrial transport | ISO | J:155856 | |||||||||
| Biological Process | GO:0006811 | monoatomic ion transport | IEA | J:60000 | |||||||||
| Biological Process | GO:0120162 | positive regulation of cold-induced thermogenesis | IMP | J:244963 | |||||||||
| Biological Process | GO:1902600 | proton transmembrane transport | IDA | J:189069 | |||||||||
| Biological Process | GO:1902600 | proton transmembrane transport | ISO | J:164563 | |||||||||
| Biological Process | GO:1902600 | proton transmembrane transport | ISO | J:155856 | |||||||||
| Biological Process | GO:1903426 | regulation of reactive oxygen species biosynthetic process | IMP | J:165368 | |||||||||
| Biological Process | GO:0006357 | regulation of transcription by RNA polymerase II | IDA | J:126723 | |||||||||
| Biological Process | GO:0009409 | response to cold | IBA | J:265628 | |||||||||
| Biological Process | GO:0031667 | response to nutrient levels | IMP | J:146646 | |||||||||
| Biological Process | GO:0009266 | response to temperature stimulus | IMP | J:146646 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 05/07/2024 MGI 6.23 |

|
|
|
||