|
Symbol Name ID |
Scd1
stearoyl-Coenzyme A desaturase 1 MGI:98239 |
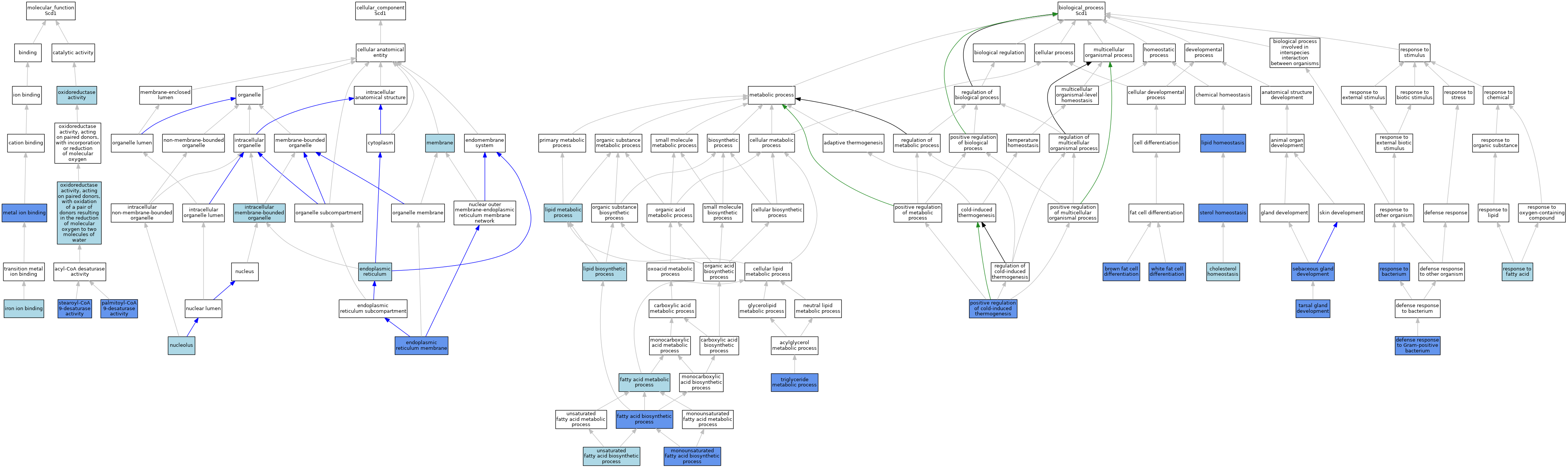
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0005506 | iron ion binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0005506 | iron ion binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0005506 | iron ion binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:0046872 | metal ion binding | IDA | J:223020 | |||||||||
| Molecular Function | GO:0016491 | oxidoreductase activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0016491 | oxidoreductase activity | ISO | J:155856 | |||||||||
| Molecular Function | GO:0016717 | oxidoreductase activity, acting on paired donors, with oxidation of a pair of donors resulting in the reduction of molecular oxygen to two molecules of water | IEA | J:72247 | |||||||||
| Molecular Function | GO:0032896 | palmitoyl-CoA 9-desaturase activity | IDA | J:223020 | |||||||||
| Molecular Function | GO:0032896 | palmitoyl-CoA 9-desaturase activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0032896 | palmitoyl-CoA 9-desaturase activity | IDA | J:108167 | |||||||||
| Molecular Function | GO:0032896 | palmitoyl-CoA 9-desaturase activity | IGI | J:108167 | |||||||||
| Molecular Function | GO:0004768 | stearoyl-CoA 9-desaturase activity | ISO | J:155856 | |||||||||
| Molecular Function | GO:0004768 | stearoyl-CoA 9-desaturase activity | IDA | J:223020 | |||||||||
| Molecular Function | GO:0004768 | stearoyl-CoA 9-desaturase activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0004768 | stearoyl-CoA 9-desaturase activity | IDA | J:108167 | |||||||||
| Molecular Function | GO:0004768 | stearoyl-CoA 9-desaturase activity | IGI | J:108167 | |||||||||
| Molecular Function | GO:0004768 | stearoyl-CoA 9-desaturase activity | IDA | J:108167 | |||||||||
| Cellular Component | GO:0005783 | endoplasmic reticulum | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005789 | endoplasmic reticulum membrane | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005789 | endoplasmic reticulum membrane | IDA | J:210917 | |||||||||
| Cellular Component | GO:0005789 | endoplasmic reticulum membrane | IBA | J:265628 | |||||||||
| Cellular Component | GO:0043231 | intracellular membrane-bounded organelle | ISO | J:155856 | |||||||||
| Cellular Component | GO:0016020 | membrane | ISO | J:164563 | |||||||||
| Cellular Component | GO:0016020 | membrane | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005730 | nucleolus | ISO | J:164563 | |||||||||
| Biological Process | GO:0050873 | brown fat cell differentiation | IDA | J:137608 | |||||||||
| Biological Process | GO:0042632 | cholesterol homeostasis | TAS | J:65994 | |||||||||
| Biological Process | GO:0050830 | defense response to Gram-positive bacterium | IMP | J:100424 | |||||||||
| Biological Process | GO:0050830 | defense response to Gram-positive bacterium | IMP | J:100424 | |||||||||
| Biological Process | GO:0006633 | fatty acid biosynthetic process | IMP | J:85276 | |||||||||
| Biological Process | GO:0006631 | fatty acid metabolic process | IEA | J:60000 | |||||||||
| Biological Process | GO:0008610 | lipid biosynthetic process | TAS | J:225402 | |||||||||
| Biological Process | GO:0055088 | lipid homeostasis | IMP | J:100424 | |||||||||
| Biological Process | GO:0006629 | lipid metabolic process | IEA | J:72247 | |||||||||
| Biological Process | GO:0006629 | lipid metabolic process | IEA | J:60000 | |||||||||
| Biological Process | GO:1903966 | monounsaturated fatty acid biosynthetic process | IMP | J:64731 | |||||||||
| Biological Process | GO:1903966 | monounsaturated fatty acid biosynthetic process | IMP | J:223020 | |||||||||
| Biological Process | GO:1903966 | monounsaturated fatty acid biosynthetic process | IGI | J:108167 | |||||||||
| Biological Process | GO:1903966 | monounsaturated fatty acid biosynthetic process | IDA | J:108167 | |||||||||
| Biological Process | GO:0120162 | positive regulation of cold-induced thermogenesis | IMP | J:152631 | |||||||||
| Biological Process | GO:0120162 | positive regulation of cold-induced thermogenesis | IMP | J:93618 | |||||||||
| Biological Process | GO:0009617 | response to bacterium | IEP | J:190322 | |||||||||
| Biological Process | GO:0070542 | response to fatty acid | ISO | J:155856 | |||||||||
| Biological Process | GO:0070542 | response to fatty acid | IBA | J:265628 | |||||||||
| Biological Process | GO:0048733 | sebaceous gland development | IMP | J:62699 | |||||||||
| Biological Process | GO:0055092 | sterol homeostasis | IMP | J:62699 | |||||||||
| Biological Process | GO:1903699 | tarsal gland development | IMP | J:62699 | |||||||||
| Biological Process | GO:0006641 | triglyceride metabolic process | IMP | J:64731 | |||||||||
| Biological Process | GO:0006636 | unsaturated fatty acid biosynthetic process | ISO | J:164563 | |||||||||
| Biological Process | GO:0006636 | unsaturated fatty acid biosynthetic process | ISO | J:155856 | |||||||||
| Biological Process | GO:0006636 | unsaturated fatty acid biosynthetic process | IBA | J:265628 | |||||||||
| Biological Process | GO:0050872 | white fat cell differentiation | IDA | J:137608 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/16/2024 MGI 6.23 |

|
|
|
||