|
Symbol Name ID |
Ptpn5
protein tyrosine phosphatase, non-receptor type 5 MGI:97807 |
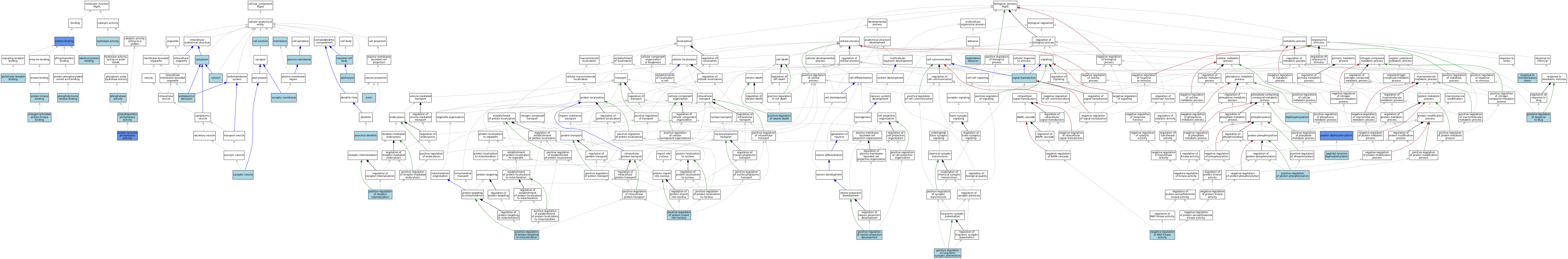
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0035254 | glutamate receptor binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0016787 | hydrolase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0042802 | identical protein binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0051019 | mitogen-activated protein kinase binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0016791 | phosphatase activity | IEA | J:72247 | |||||||||
| Molecular Function | GO:0004721 | phosphoprotein phosphatase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0001784 | phosphotyrosine residue binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:245292 | |||||||||
| Molecular Function | GO:0019901 | protein kinase binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0019901 | protein kinase binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:0004725 | protein tyrosine phosphatase activity | ISO | J:155856 | |||||||||
| Molecular Function | GO:0004725 | protein tyrosine phosphatase activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0004725 | protein tyrosine phosphatase activity | IMP | J:101438 | |||||||||
| Cellular Component | GO:0030424 | axon | ISO | J:155856 | |||||||||
| Cellular Component | GO:0030054 | cell junction | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005829 | cytosol | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005829 | cytosol | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005783 | endoplasmic reticulum | ISO | J:155856 | |||||||||
| Cellular Component | GO:0016020 | membrane | ISO | J:155856 | |||||||||
| Cellular Component | GO:0043025 | neuronal cell body | ISO | J:155856 | |||||||||
| Cellular Component | GO:0043204 | perikaryon | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | IBA | J:265628 | |||||||||
| Cellular Component | GO:1990635 | proximal dendrite | ISO | J:155856 | |||||||||
| Cellular Component | GO:0097060 | synaptic membrane | ISO | J:155856 | |||||||||
| Cellular Component | GO:0008021 | synaptic vesicle | ISO | J:155856 | |||||||||
| Biological Process | GO:0016311 | dephosphorylation | IEA | J:72247 | |||||||||
| Biological Process | GO:0035640 | exploration behavior | ISO | J:155856 | |||||||||
| Biological Process | GO:0043407 | negative regulation of MAP kinase activity | ISO | J:155856 | |||||||||
| Biological Process | GO:0035335 | peptidyl-tyrosine dephosphorylation | ISO | J:155856 | |||||||||
| Biological Process | GO:0035335 | peptidyl-tyrosine dephosphorylation | IBA | J:265628 | |||||||||
| Biological Process | GO:1900273 | positive regulation of long-term synaptic potentiation | ISO | J:155856 | |||||||||
| Biological Process | GO:1901216 | positive regulation of neuron death | ISO | J:155856 | |||||||||
| Biological Process | GO:0010976 | positive regulation of neuron projection development | ISO | J:155856 | |||||||||
| Biological Process | GO:0042307 | positive regulation of protein import into nucleus | ISO | J:155856 | |||||||||
| Biological Process | GO:0001934 | positive regulation of protein phosphorylation | ISO | J:155856 | |||||||||
| Biological Process | GO:1903955 | positive regulation of protein targeting to mitochondrion | ISO | J:155856 | |||||||||
| Biological Process | GO:0002092 | positive regulation of receptor internalization | ISO | J:155856 | |||||||||
| Biological Process | GO:2001025 | positive regulation of response to drug | ISO | J:155856 | |||||||||
| Biological Process | GO:0006470 | protein dephosphorylation | IMP | J:101438 | |||||||||
| Biological Process | GO:0035902 | response to immobilization stress | ISO | J:155856 | |||||||||
| Biological Process | GO:0007165 | signal transduction | IBA | J:265628 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/30/2024 MGI 6.23 |

|
|
|
||