|
Symbol Name ID |
Plp1
proteolipid protein (myelin) 1 MGI:97623 |
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
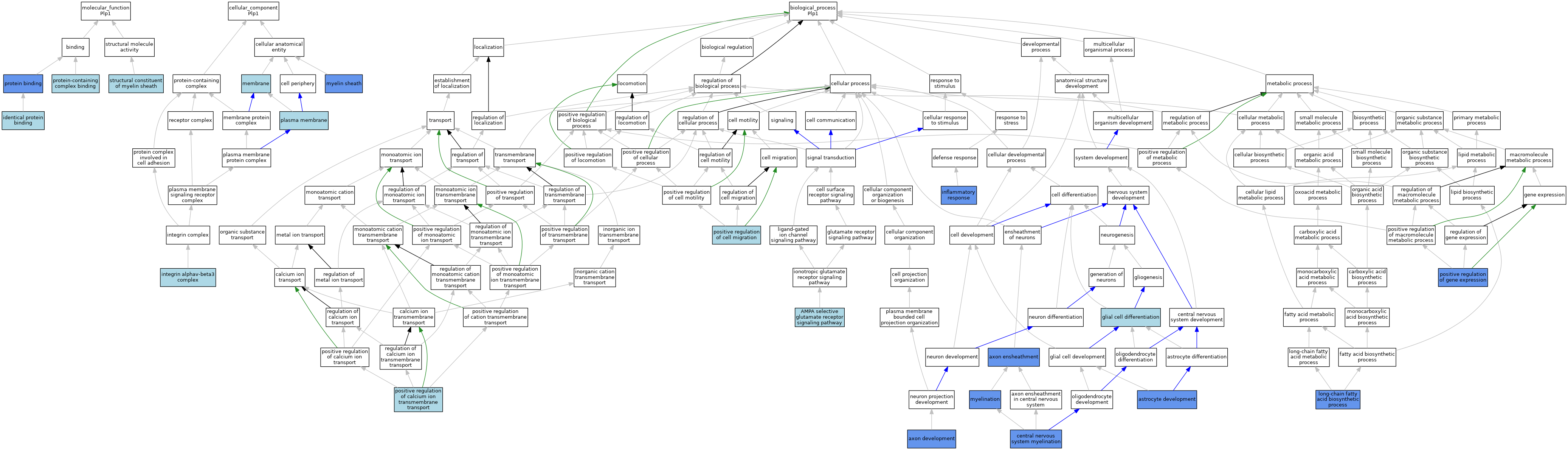
| Molecular Function | GO:0042802 | identical protein binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:93976 | |||||||||
| Molecular Function | GO:0044877 | protein-containing complex binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0019911 | structural constituent of myelin sheath | ISO | J:155856 | |||||||||
| Molecular Function | GO:0019911 | structural constituent of myelin sheath | IBA | J:265628 | |||||||||
| Cellular Component | GO:0034683 | integrin alphav-beta3 complex | ISO | J:155856 | |||||||||
| Cellular Component | GO:0016020 | membrane | IEA | J:72247 | |||||||||
| Cellular Component | GO:0016020 | membrane | IEA | J:60000 | |||||||||
| Cellular Component | GO:0043209 | myelin sheath | HDA | J:145263 | |||||||||
| Cellular Component | GO:0043209 | myelin sheath | ISO | J:155856 | |||||||||
| Cellular Component | GO:0043209 | myelin sheath | IBA | J:265628 | |||||||||
| Cellular Component | GO:0043209 | myelin sheath | IDA | J:167272 | |||||||||
| Cellular Component | GO:0043209 | myelin sheath | IDA | J:10575 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | IBA | J:265628 | |||||||||
| Biological Process | GO:0098990 | AMPA selective glutamate receptor signaling pathway | ISO | J:155856 | |||||||||
| Biological Process | GO:0014002 | astrocyte development | IGI | J:201143 | |||||||||
| Biological Process | GO:0061564 | axon development | IBA | J:265628 | |||||||||
| Biological Process | GO:0061564 | axon development | IGI | J:201143 | |||||||||
| Biological Process | GO:0008366 | axon ensheathment | IMP | J:13141 | |||||||||
| Biological Process | GO:0022010 | central nervous system myelination | IBA | J:265628 | |||||||||
| Biological Process | GO:0022010 | central nervous system myelination | IMP | J:10575 | |||||||||
| Biological Process | GO:0010001 | glial cell differentiation | ISO | J:155856 | |||||||||
| Biological Process | GO:0006954 | inflammatory response | IGI | J:201143 | |||||||||
| Biological Process | GO:0042759 | long-chain fatty acid biosynthetic process | IMP | J:106960 | |||||||||
| Biological Process | GO:0042552 | myelination | ISO | J:155856 | |||||||||
| Biological Process | GO:0042552 | myelination | IGI | J:201143 | |||||||||
| Biological Process | GO:0042552 | myelination | TAS | J:77770 | |||||||||
| Biological Process | GO:0042552 | myelination | IMP | J:849 | |||||||||
| Biological Process | GO:0042552 | myelination | IMP | J:242069 | |||||||||
| Biological Process | GO:0042552 | myelination | IMP | J:106960 | |||||||||
| Biological Process | GO:1904427 | positive regulation of calcium ion transmembrane transport | ISO | J:155856 | |||||||||
| Biological Process | GO:0030335 | positive regulation of cell migration | ISO | J:155856 | |||||||||
| Biological Process | GO:0010628 | positive regulation of gene expression | IMP | J:145263 | |||||||||
| Biological Process | GO:0010628 | positive regulation of gene expression | IMP | J:849 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/16/2024 MGI 6.23 |

|
|
|
||