|
Symbol Name ID |
Plaur
plasminogen activator, urokinase receptor MGI:97612 |
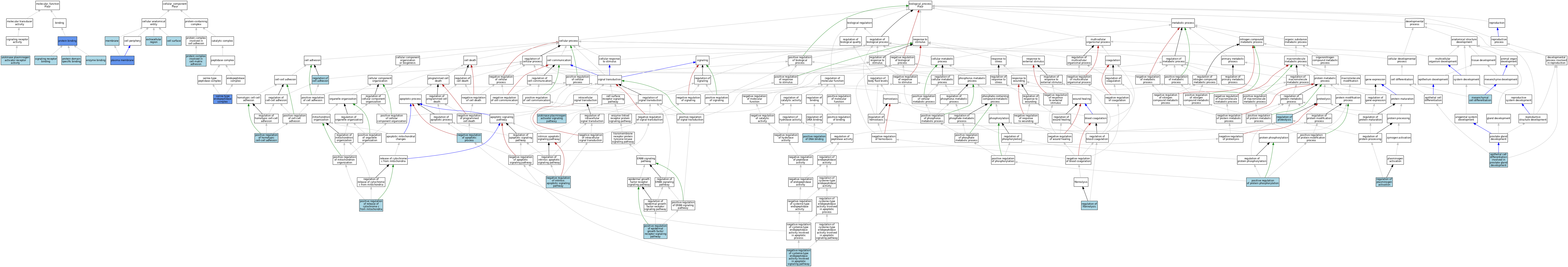
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0019899 | enzyme binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:155175 | |||||||||
| Molecular Function | GO:0019904 | protein domain specific binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0005102 | signaling receptor binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0030377 | urokinase plasminogen activator receptor activity | IEA | J:72247 | |||||||||
| Cellular Component | GO:0009986 | cell surface | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005576 | extracellular region | IEA | J:60000 | |||||||||
| Cellular Component | GO:0016020 | membrane | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | IDA | J:157880 | |||||||||
| Cellular Component | GO:0098637 | protein complex involved in cell-matrix adhesion | ISO | J:164563 | |||||||||
| Cellular Component | GO:1905370 | serine-type endopeptidase complex | ISO | J:164563 | |||||||||
| Cellular Component | GO:1905370 | serine-type endopeptidase complex | IPI | J:165944 | |||||||||
| Biological Process | GO:0060742 | epithelial cell differentiation involved in prostate gland development | ISO | J:155856 | |||||||||
| Biological Process | GO:0048762 | mesenchymal cell differentiation | ISO | J:155856 | |||||||||
| Biological Process | GO:0043066 | negative regulation of apoptotic process | ISO | J:155856 | |||||||||
| Biological Process | GO:0043066 | negative regulation of apoptotic process | ISO | J:164563 | |||||||||
| Biological Process | GO:2001268 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway | ISO | J:164563 | |||||||||
| Biological Process | GO:2001243 | negative regulation of intrinsic apoptotic signaling pathway | ISO | J:164563 | |||||||||
| Biological Process | GO:0043388 | positive regulation of DNA binding | ISO | J:164563 | |||||||||
| Biological Process | GO:0045742 | positive regulation of epidermal growth factor receptor signaling pathway | ISO | J:164563 | |||||||||
| Biological Process | GO:0034112 | positive regulation of homotypic cell-cell adhesion | ISO | J:164563 | |||||||||
| Biological Process | GO:0001934 | positive regulation of protein phosphorylation | ISO | J:164563 | |||||||||
| Biological Process | GO:0090200 | positive regulation of release of cytochrome c from mitochondria | ISO | J:164563 | |||||||||
| Biological Process | GO:0030155 | regulation of cell adhesion | ISO | J:164563 | |||||||||
| Biological Process | GO:0051917 | regulation of fibrinolysis | NAS | J:320117 | |||||||||
| Biological Process | GO:0010755 | regulation of plasminogen activation | NAS | J:320117 | |||||||||
| Biological Process | GO:0030162 | regulation of proteolysis | IEA | J:72247 | |||||||||
| Biological Process | GO:0038195 | urokinase plasminogen activator signaling pathway | NAS | J:320117 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/30/2024 MGI 6.23 |

|
|
|
||