|
Symbol Name ID |
Furin
furin, paired basic amino acid cleaving enzyme MGI:97513 |
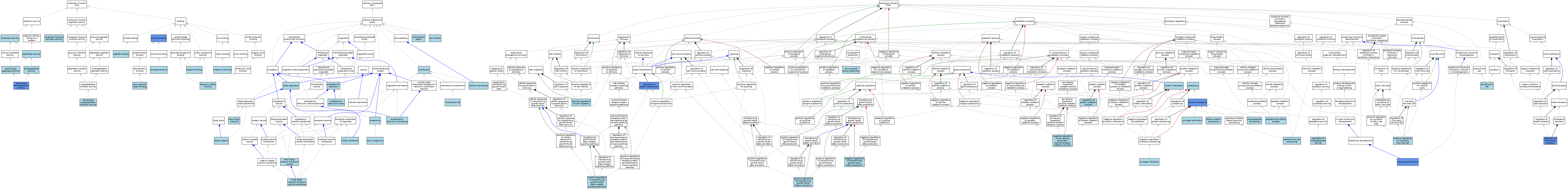
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0004175 | endopeptidase activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:1904399 | heparan sulfate binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0008201 | heparin binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0016787 | hydrolase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0046872 | metal ion binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0140677 | molecular function activator activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0048406 | nerve growth factor binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0008233 | peptidase activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0008233 | peptidase activity | ISO | J:95653 | |||||||||
| Molecular Function | GO:0008233 | peptidase activity | ISO | J:90197 | |||||||||
| Molecular Function | GO:0042277 | peptide binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0002020 | protease binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:51716 | |||||||||
| Molecular Function | GO:0004252 | serine-type endopeptidase activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0004252 | serine-type endopeptidase activity | EXP | J:81247 | |||||||||
| Molecular Function | GO:0004252 | serine-type endopeptidase activity | ISO | J:155856 | |||||||||
| Molecular Function | GO:0004252 | serine-type endopeptidase activity | TAS | Reactome:R-MMU-2484926 | |||||||||
| Molecular Function | GO:0004252 | serine-type endopeptidase activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0004867 | serine-type endopeptidase inhibitor activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0008236 | serine-type peptidase activity | ISO | J:164563 | |||||||||
| Cellular Component | GO:0009986 | cell surface | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005769 | early endosome | TAS | J:79598 | |||||||||
| Cellular Component | GO:0005783 | endoplasmic reticulum | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005788 | endoplasmic reticulum lumen | TAS | Reactome:R-MMU-181788 | |||||||||
| Cellular Component | GO:0005789 | endoplasmic reticulum membrane | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005768 | endosome | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005576 | extracellular region | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005576 | extracellular region | TAS | Reactome:R-MMU-1181148 | |||||||||
| Cellular Component | GO:0005576 | extracellular region | TAS | Reactome:R-MMU-2484926 | |||||||||
| Cellular Component | GO:0005794 | Golgi apparatus | IEA | J:60000 | |||||||||
| Cellular Component | GO:0031985 | Golgi cisterna | ISO | J:155856 | |||||||||
| Cellular Component | GO:0000139 | Golgi membrane | ISO | J:164563 | |||||||||
| Cellular Component | GO:0016020 | membrane | IBA | J:265628 | |||||||||
| Cellular Component | GO:0016020 | membrane | TAS | J:79598 | |||||||||
| Cellular Component | GO:0045121 | membrane raft | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005802 | trans-Golgi network | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005802 | trans-Golgi network | IBA | J:265628 | |||||||||
| Cellular Component | GO:0030140 | trans-Golgi network transport vesicle | ISO | J:89488 | |||||||||
| Cellular Component | GO:0012510 | trans-Golgi network transport vesicle membrane | TAS | J:79598 | |||||||||
| Biological Process | GO:0001825 | blastocyst formation | IGI | J:227651 | |||||||||
| Biological Process | GO:0090472 | dibasic protein processing | ISO | J:164563 | |||||||||
| Biological Process | GO:0032804 | negative regulation of low-density lipoprotein particle receptor catabolic process | ISO | J:164563 | |||||||||
| Biological Process | GO:0032911 | negative regulation of transforming growth factor beta1 production | ISO | J:164563 | |||||||||
| Biological Process | GO:0032902 | nerve growth factor production | ISO | J:164563 | |||||||||
| Biological Process | GO:0043043 | peptide biosynthetic process | ISO | J:164563 | |||||||||
| Biological Process | GO:0016486 | peptide hormone processing | ISO | J:164563 | |||||||||
| Biological Process | GO:0016486 | peptide hormone processing | IBA | J:265628 | |||||||||
| Biological Process | GO:0030335 | positive regulation of cell migration | ISO | J:155856 | |||||||||
| Biological Process | GO:0030511 | positive regulation of transforming growth factor beta receptor signaling pathway | ISO | J:155856 | |||||||||
| Biological Process | GO:1901394 | positive regulation of transforming growth factor beta1 activation | ISO | J:164563 | |||||||||
| Biological Process | GO:0046598 | positive regulation of viral entry into host cell | ISO | J:164563 | |||||||||
| Biological Process | GO:0051604 | protein maturation | ISO | J:164563 | |||||||||
| Biological Process | GO:0016485 | protein processing | ISO | J:164563 | |||||||||
| Biological Process | GO:0016485 | protein processing | ISO | J:155856 | |||||||||
| Biological Process | GO:0016485 | protein processing | IMP | J:203527 | |||||||||
| Biological Process | GO:0016485 | protein processing | IMP | J:169483 | |||||||||
| Biological Process | GO:0016485 | protein processing | ISO | J:95653 | |||||||||
| Biological Process | GO:0016485 | protein processing | IDA | J:163721 | |||||||||
| Biological Process | GO:0006508 | proteolysis | IEA | J:72247 | |||||||||
| Biological Process | GO:0006508 | proteolysis | IEA | J:60000 | |||||||||
| Biological Process | GO:0032374 | regulation of cholesterol transport | IMP | J:169483 | |||||||||
| Biological Process | GO:0052548 | regulation of endopeptidase activity | ISO | J:164563 | |||||||||
| Biological Process | GO:0042176 | regulation of protein catabolic process | ISO | J:164563 | |||||||||
| Biological Process | GO:0009966 | regulation of signal transduction | IDA | J:127541 | |||||||||
| Biological Process | GO:0032940 | secretion by cell | ISO | J:164563 | |||||||||
| Biological Process | GO:0006465 | signal peptide processing | ISO | J:164563 | |||||||||
| Biological Process | GO:0031638 | zymogen activation | ISO | J:164563 | |||||||||
| Biological Process | GO:0097341 | zymogen inhibition | ISO | J:164563 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/16/2024 MGI 6.23 |

|
|
|
||