|
Symbol Name ID |
Mmut
methylmalonyl-Coenzyme A mutase MGI:97239 |
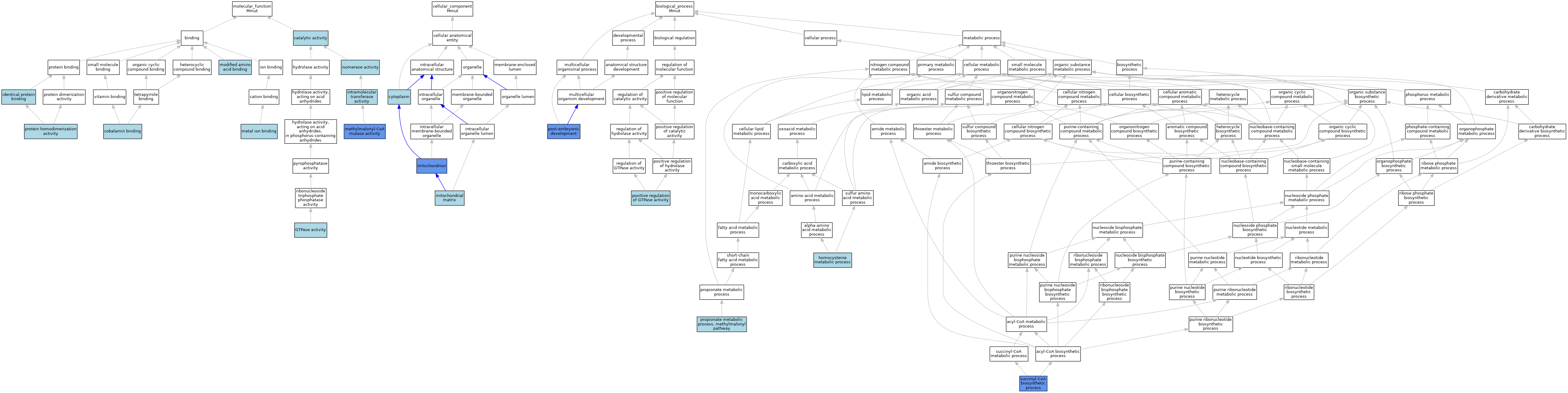
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0003824 | catalytic activity | IEA | J:72247 | |||||||||
| Molecular Function | GO:0031419 | cobalamin binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0031419 | cobalamin binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:0003924 | GTPase activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0042802 | identical protein binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0016866 | intramolecular transferase activity | IEA | J:72247 | |||||||||
| Molecular Function | GO:0016853 | isomerase activity | IEA | J:72247 | |||||||||
| Molecular Function | GO:0016853 | isomerase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0046872 | metal ion binding | IEA | J:72247 | |||||||||
| Molecular Function | GO:0046872 | metal ion binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0004494 | methylmalonyl-CoA mutase activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0004494 | methylmalonyl-CoA mutase activity | IDA | J:10823 | |||||||||
| Molecular Function | GO:0004494 | methylmalonyl-CoA mutase activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0004494 | methylmalonyl-CoA mutase activity | IDA | J:16322 | |||||||||
| Molecular Function | GO:0004494 | methylmalonyl-CoA mutase activity | IDA | J:87081 | |||||||||
| Molecular Function | GO:0072341 | modified amino acid binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0042803 | protein homodimerization activity | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005759 | mitochondrial matrix | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005739 | mitochondrion | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005739 | mitochondrion | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005739 | mitochondrion | HDA | J:86816 | |||||||||
| Cellular Component | GO:0005739 | mitochondrion | HDA | J:151002 | |||||||||
| Cellular Component | GO:0005739 | mitochondrion | HDA | J:86816 | |||||||||
| Cellular Component | GO:0005739 | mitochondrion | HDA | J:86816 | |||||||||
| Biological Process | GO:0050667 | homocysteine metabolic process | ISO | J:164563 | |||||||||
| Biological Process | GO:0043547 | positive regulation of GTPase activity | ISO | J:164563 | |||||||||
| Biological Process | GO:0009791 | post-embryonic development | IMP | J:87081 | |||||||||
| Biological Process | GO:0019678 | propionate metabolic process, methylmalonyl pathway | IBA | J:265628 | |||||||||
| Biological Process | GO:1901290 | succinyl-CoA biosynthetic process | IDA | J:16322 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 05/07/2024 MGI 6.23 |

|
|
|
||