|
Symbol Name ID |
Il6st
interleukin 6 signal transducer MGI:96560 |
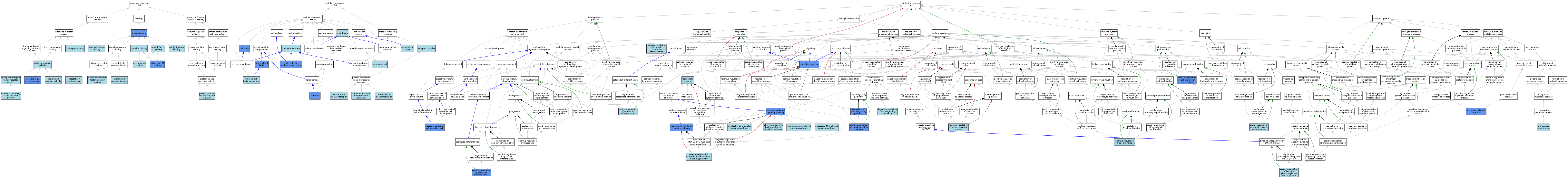
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0004897 | ciliary neurotrophic factor receptor activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0005127 | ciliary neurotrophic factor receptor binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0015026 | coreceptor activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0019955 | cytokine binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:0004896 | cytokine receptor activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0019838 | growth factor binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0019838 | growth factor binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0042802 | identical protein binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0019970 | interleukin-11 binding | IDA | J:21233 | |||||||||
| Molecular Function | GO:0004921 | interleukin-11 receptor activity | IDA | J:21233 | |||||||||
| Molecular Function | GO:0019981 | interleukin-6 binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0019981 | interleukin-6 binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:0004915 | interleukin-6 receptor activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0004915 | interleukin-6 receptor activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0005138 | interleukin-6 receptor binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0004923 | leukemia inhibitory factor receptor activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0004924 | oncostatin-M receptor activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:304576 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:201263 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:105174 | |||||||||
| Molecular Function | GO:0030296 | protein tyrosine kinase activator activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0030296 | protein tyrosine kinase activator activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0097110 | scaffold protein binding | ISO | J:164563 | |||||||||
| Cellular Component | GO:0044297 | cell body | IDA | J:74068 | |||||||||
| Cellular Component | GO:0070110 | ciliary neurotrophic factor receptor complex | ISO | J:164563 | |||||||||
| Cellular Component | GO:0030425 | dendrite | IDA | J:74068 | |||||||||
| Cellular Component | GO:0009897 | external side of plasma membrane | IBA | J:265628 | |||||||||
| Cellular Component | GO:0009897 | external side of plasma membrane | IDA | J:68367 | |||||||||
| Cellular Component | GO:0005615 | extracellular space | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005896 | interleukin-6 receptor complex | ISO | J:164563 | |||||||||
| Cellular Component | GO:0016020 | membrane | IEA | J:72247 | |||||||||
| Cellular Component | GO:0016020 | membrane | IEA | J:60000 | |||||||||
| Cellular Component | GO:0045121 | membrane raft | ISO | J:164563 | |||||||||
| Cellular Component | GO:0043025 | neuronal cell body | IDA | J:74068 | |||||||||
| Cellular Component | GO:0032809 | neuronal cell body membrane | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005900 | oncostatin-M receptor complex | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | ISO | J:164563 | |||||||||
| Cellular Component | GO:0043235 | receptor complex | IBA | J:265628 | |||||||||
| Biological Process | GO:0070120 | ciliary neurotrophic factor-mediated signaling pathway | ISO | J:164563 | |||||||||
| Biological Process | GO:0019221 | cytokine-mediated signaling pathway | ISO | J:164563 | |||||||||
| Biological Process | GO:0019221 | cytokine-mediated signaling pathway | IBA | J:265628 | |||||||||
| Biological Process | GO:0019221 | cytokine-mediated signaling pathway | IDA | J:21233 | |||||||||
| Biological Process | GO:0005977 | glycogen metabolic process | IMP | J:54660 | |||||||||
| Biological Process | GO:0038154 | interleukin-11-mediated signaling pathway | ISO | J:164563 | |||||||||
| Biological Process | GO:0070106 | interleukin-27-mediated signaling pathway | ISO | J:164563 | |||||||||
| Biological Process | GO:0070102 | interleukin-6-mediated signaling pathway | IDA | J:219844 | |||||||||
| Biological Process | GO:0070102 | interleukin-6-mediated signaling pathway | ISO | J:155856 | |||||||||
| Biological Process | GO:0070102 | interleukin-6-mediated signaling pathway | ISO | J:164563 | |||||||||
| Biological Process | GO:0060576 | intestinal epithelial cell development | ISO | J:164563 | |||||||||
| Biological Process | GO:0060576 | intestinal epithelial cell development | IDA | J:219844 | |||||||||
| Biological Process | GO:0048861 | leukemia inhibitory factor signaling pathway | ISO | J:164563 | |||||||||
| Biological Process | GO:0051481 | negative regulation of cytosolic calcium ion concentration | ISO | J:155856 | |||||||||
| Biological Process | GO:0070104 | negative regulation of interleukin-6-mediated signaling pathway | ISO | J:164563 | |||||||||
| Biological Process | GO:0043524 | negative regulation of neuron apoptotic process | ISO | J:164563 | |||||||||
| Biological Process | GO:0038165 | oncostatin-M-mediated signaling pathway | ISO | J:164563 | |||||||||
| Biological Process | GO:0048711 | positive regulation of astrocyte differentiation | IMP | J:57039 | |||||||||
| Biological Process | GO:0008284 | positive regulation of cell population proliferation | ISO | J:164563 | |||||||||
| Biological Process | GO:0008284 | positive regulation of cell population proliferation | IGI | J:89246 | |||||||||
| Biological Process | GO:0045747 | positive regulation of Notch signaling pathway | IDA | J:219844 | |||||||||
| Biological Process | GO:0045747 | positive regulation of Notch signaling pathway | ISO | J:164563 | |||||||||
| Biological Process | GO:0045669 | positive regulation of osteoblast differentiation | ISO | J:164563 | |||||||||
| Biological Process | GO:0014911 | positive regulation of smooth muscle cell migration | ISO | J:155856 | |||||||||
| Biological Process | GO:0042102 | positive regulation of T cell proliferation | ISO | J:164563 | |||||||||
| Biological Process | GO:0042531 | positive regulation of tyrosine phosphorylation of STAT protein | ISO | J:164563 | |||||||||
| Biological Process | GO:0008593 | regulation of Notch signaling pathway | IDA | J:82269 | |||||||||
| Biological Process | GO:0034097 | response to cytokine | ISO | J:164563 | |||||||||
| Biological Process | GO:0007165 | signal transduction | IDA | J:79290 | |||||||||
| Biological Process | GO:0006642 | triglyceride mobilization | ISO | J:155856 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/16/2024 MGI 6.23 |

|
|
|
||