|
Symbol Name ID |
Lipc
lipase, hepatic MGI:96216 |
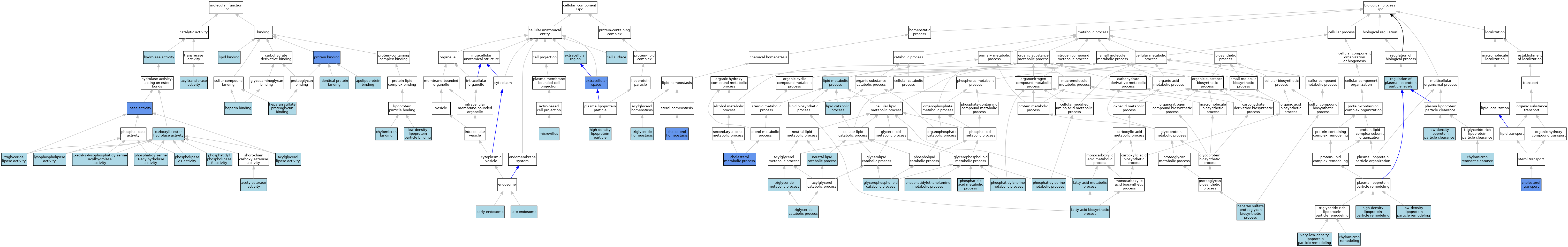
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0052740 | 1-acyl-2-lysophosphatidylserine acylhydrolase activity | IEA | J:72245 | |||||||||
| Molecular Function | GO:0008126 | acetylesterase activity | ISO | J:155856 | |||||||||
| Molecular Function | GO:0047372 | acylglycerol lipase activity | ISO | J:155856 | |||||||||
| Molecular Function | GO:0016746 | acyltransferase activity | ISO | J:155856 | |||||||||
| Molecular Function | GO:0034185 | apolipoprotein binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0034185 | apolipoprotein binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:0052689 | carboxylic ester hydrolase activity | IEA | J:72247 | |||||||||
| Molecular Function | GO:0035478 | chylomicron binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0043395 | heparan sulfate proteoglycan binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0008201 | heparin binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0016787 | hydrolase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0042802 | identical protein binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0016298 | lipase activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0016298 | lipase activity | IDA | J:100492 | |||||||||
| Molecular Function | GO:0016298 | lipase activity | IDA | J:85532 | |||||||||
| Molecular Function | GO:0008289 | lipid binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0030169 | low-density lipoprotein particle binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0004622 | lysophospholipase activity | ISO | J:155856 | |||||||||
| Molecular Function | GO:0102545 | phosphatidyl phospholipase B activity | IEA | J:72245 | |||||||||
| Molecular Function | GO:0052739 | phosphatidylserine 1-acylhydrolase activity | IEA | J:72245 | |||||||||
| Molecular Function | GO:0008970 | phospholipase A1 activity | ISO | J:155856 | |||||||||
| Molecular Function | GO:0008970 | phospholipase A1 activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:156963 | |||||||||
| Molecular Function | GO:0004806 | triglyceride lipase activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0004806 | triglyceride lipase activity | ISO | J:155856 | |||||||||
| Cellular Component | GO:0009986 | cell surface | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005769 | early endosome | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005576 | extracellular region | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005615 | extracellular space | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005615 | extracellular space | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005615 | extracellular space | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005615 | extracellular space | IDA | J:100492 | |||||||||
| Cellular Component | GO:0034364 | high-density lipoprotein particle | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005770 | late endosome | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005902 | microvillus | ISO | J:155856 | |||||||||
| Biological Process | GO:0042632 | cholesterol homeostasis | ISO | J:164563 | |||||||||
| Biological Process | GO:0042632 | cholesterol homeostasis | IGI | J:55745 | |||||||||
| Biological Process | GO:0008203 | cholesterol metabolic process | IGI | J:55745 | |||||||||
| Biological Process | GO:0030301 | cholesterol transport | IMP | J:35143 | |||||||||
| Biological Process | GO:0034382 | chylomicron remnant clearance | ISO | J:155856 | |||||||||
| Biological Process | GO:0034371 | chylomicron remodeling | ISO | J:155856 | |||||||||
| Biological Process | GO:0006633 | fatty acid biosynthetic process | ISO | J:164563 | |||||||||
| Biological Process | GO:0006631 | fatty acid metabolic process | ISO | J:155856 | |||||||||
| Biological Process | GO:0046475 | glycerophospholipid catabolic process | ISO | J:155856 | |||||||||
| Biological Process | GO:0015012 | heparan sulfate proteoglycan biosynthetic process | ISO | J:155856 | |||||||||
| Biological Process | GO:0034375 | high-density lipoprotein particle remodeling | ISO | J:164563 | |||||||||
| Biological Process | GO:0034375 | high-density lipoprotein particle remodeling | ISO | J:155856 | |||||||||
| Biological Process | GO:0016042 | lipid catabolic process | IBA | J:265628 | |||||||||
| Biological Process | GO:0006629 | lipid metabolic process | IEA | J:60000 | |||||||||
| Biological Process | GO:0006629 | lipid metabolic process | IEA | J:72247 | |||||||||
| Biological Process | GO:0034383 | low-density lipoprotein particle clearance | ISO | J:155856 | |||||||||
| Biological Process | GO:0034374 | low-density lipoprotein particle remodeling | ISO | J:164563 | |||||||||
| Biological Process | GO:0034374 | low-density lipoprotein particle remodeling | ISO | J:155856 | |||||||||
| Biological Process | GO:0046461 | neutral lipid catabolic process | ISO | J:155856 | |||||||||
| Biological Process | GO:0046473 | phosphatidic acid metabolic process | ISO | J:155856 | |||||||||
| Biological Process | GO:0046470 | phosphatidylcholine metabolic process | ISO | J:155856 | |||||||||
| Biological Process | GO:0046337 | phosphatidylethanolamine metabolic process | ISO | J:155856 | |||||||||
| Biological Process | GO:0006658 | phosphatidylserine metabolic process | ISO | J:155856 | |||||||||
| Biological Process | GO:0097006 | regulation of plasma lipoprotein particle levels | ISO | J:155856 | |||||||||
| Biological Process | GO:0019433 | triglyceride catabolic process | ISO | J:164563 | |||||||||
| Biological Process | GO:0070328 | triglyceride homeostasis | ISO | J:164563 | |||||||||
| Biological Process | GO:0006641 | triglyceride metabolic process | ISO | J:155856 | |||||||||
| Biological Process | GO:0034372 | very-low-density lipoprotein particle remodeling | ISO | J:155856 | |||||||||
| Biological Process | GO:0034372 | very-low-density lipoprotein particle remodeling | ISO | J:164563 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/16/2024 MGI 6.23 |

|
|
|
||