|
Symbol Name ID |
Pdia3
protein disulfide isomerase associated 3 MGI:95834 |
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
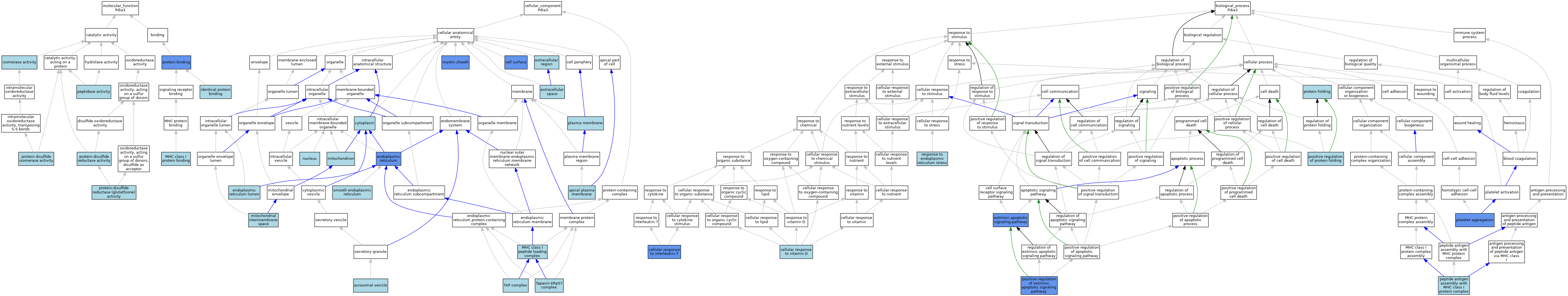
| Molecular Function | GO:0042802 | identical protein binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0016853 | isomerase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0042288 | MHC class I protein binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0008233 | peptidase activity | ISO | J:155856 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:167004 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:289677 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:198051 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:198051 | |||||||||
| Molecular Function | GO:0003756 | protein disulfide isomerase activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0019153 | protein-disulfide reductase (glutathione) activity | ISO | J:155856 | |||||||||
| Molecular Function | GO:0015035 | protein-disulfide reductase activity | ISO | J:164563 | |||||||||
| Cellular Component | GO:0001669 | acrosomal vesicle | ISO | J:155856 | |||||||||
| Cellular Component | GO:0016324 | apical plasma membrane | ISO | J:155856 | |||||||||
| Cellular Component | GO:0009986 | cell surface | ISO | J:155856 | |||||||||
| Cellular Component | GO:0009986 | cell surface | IBA | J:265628 | |||||||||
| Cellular Component | GO:0009986 | cell surface | IDA | J:182119 | |||||||||
| Cellular Component | GO:0009986 | cell surface | ISO | J:73065 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005783 | endoplasmic reticulum | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005783 | endoplasmic reticulum | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005783 | endoplasmic reticulum | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005783 | endoplasmic reticulum | IDA | J:46305 | |||||||||
| Cellular Component | GO:0005788 | endoplasmic reticulum lumen | TAS | Reactome:R-MMU-983160 | |||||||||
| Cellular Component | GO:0005576 | extracellular region | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005615 | extracellular space | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005615 | extracellular space | ISO | J:164563 | |||||||||
| Cellular Component | GO:0042824 | MHC class I peptide loading complex | ISO | J:155856 | |||||||||
| Cellular Component | GO:0042824 | MHC class I peptide loading complex | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005758 | mitochondrial intermembrane space | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005739 | mitochondrion | ISO | J:155856 | |||||||||
| Cellular Component | GO:0043209 | myelin sheath | HDA | J:145263 | |||||||||
| Cellular Component | GO:0005634 | nucleus | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005790 | smooth endoplasmic reticulum | ISO | J:155856 | |||||||||
| Cellular Component | GO:0042825 | TAP complex | ISO | J:155856 | |||||||||
| Cellular Component | GO:0061779 | Tapasin-ERp57 complex | ISO | J:164563 | |||||||||
| Biological Process | GO:0098761 | cellular response to interleukin-7 | IDA | J:260356 | |||||||||
| Biological Process | GO:0071305 | cellular response to vitamin D | ISO | J:155856 | |||||||||
| Biological Process | GO:0097191 | extrinsic apoptotic signaling pathway | IMP | J:98374 | |||||||||
| Biological Process | GO:0097191 | extrinsic apoptotic signaling pathway | IMP | J:98374 | |||||||||
| Biological Process | GO:0002502 | peptide antigen assembly with MHC class I protein complex | ISO | J:164563 | |||||||||
| Biological Process | GO:0070527 | platelet aggregation | IMP | J:244748 | |||||||||
| Biological Process | GO:2001238 | positive regulation of extrinsic apoptotic signaling pathway | IMP | J:98374 | |||||||||
| Biological Process | GO:1903334 | positive regulation of protein folding | ISO | J:155856 | |||||||||
| Biological Process | GO:0006457 | protein folding | IBA | J:265628 | |||||||||
| Biological Process | GO:0034976 | response to endoplasmic reticulum stress | IBA | J:265628 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/30/2024 MGI 6.23 |

|
|
|
||