|
Symbol Name ID |
Ghr
growth hormone receptor MGI:95708 |
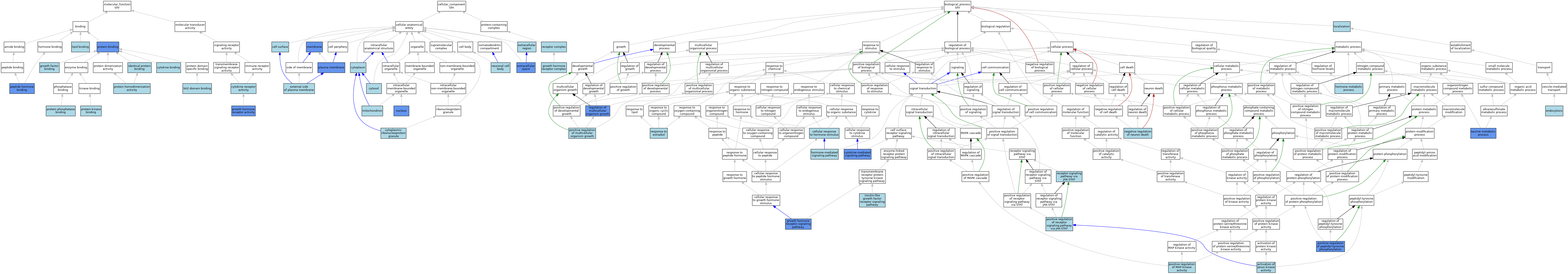
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0019955 | cytokine binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:0004896 | cytokine receptor activity | IEA | J:72247 | |||||||||
| Molecular Function | GO:0019838 | growth factor binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0019838 | growth factor binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:0004903 | growth hormone receptor activity | ISO | J:155856 | |||||||||
| Molecular Function | GO:0004903 | growth hormone receptor activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0004903 | growth hormone receptor activity | IDA | J:7060 | |||||||||
| Molecular Function | GO:0042802 | identical protein binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0008289 | lipid binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0017046 | peptide hormone binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0017046 | peptide hormone binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:0017046 | peptide hormone binding | IPI | J:58419 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:34907 | |||||||||
| Molecular Function | GO:0042803 | protein homodimerization activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0019901 | protein kinase binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0019903 | protein phosphatase binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0042169 | SH2 domain binding | ISO | J:155856 | |||||||||
| Cellular Component | GO:0009986 | cell surface | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | ISO | J:155856 | |||||||||
| Cellular Component | GO:0036464 | cytoplasmic ribonucleoprotein granule | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005829 | cytosol | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005829 | cytosol | IBA | J:265628 | |||||||||
| Cellular Component | GO:0009897 | external side of plasma membrane | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005576 | extracellular region | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005576 | extracellular region | TAS | Reactome:R-MMU-1168790 | |||||||||
| Cellular Component | GO:0005615 | extracellular space | HDA | J:221550 | |||||||||
| Cellular Component | GO:0005615 | extracellular space | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005615 | extracellular space | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005615 | extracellular space | ISO | J:155856 | |||||||||
| Cellular Component | GO:0070195 | growth hormone receptor complex | ISO | J:164563 | |||||||||
| Cellular Component | GO:0070195 | growth hormone receptor complex | IBA | J:265628 | |||||||||
| Cellular Component | GO:0016020 | membrane | ISO | J:164563 | |||||||||
| Cellular Component | GO:0016020 | membrane | IDA | J:74664 | |||||||||
| Cellular Component | GO:0005739 | mitochondrion | ISO | J:155856 | |||||||||
| Cellular Component | GO:0043025 | neuronal cell body | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005634 | nucleus | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IDA | J:40378 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IDA | J:40378 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IDA | J:40378 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | TAS | Reactome:R-MMU-1168790 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | TAS | Reactome:R-MMU-1168910 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | TAS | Reactome:R-MMU-1168923 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | TAS | Reactome:R-MMU-1168927 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | TAS | Reactome:R-MMU-1169142 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | TAS | Reactome:R-MMU-1169194 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | TAS | Reactome:R-MMU-1169206 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | TAS | Reactome:R-MMU-1169229 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | TAS | Reactome:R-MMU-1169240 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | TAS | Reactome:R-MMU-1169250 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | IDA | J:40378 | |||||||||
| Cellular Component | GO:0043235 | receptor complex | ISO | J:164563 | |||||||||
| Biological Process | GO:0042976 | activation of Janus kinase activity | ISO | J:155856 | |||||||||
| Biological Process | GO:0032870 | cellular response to hormone stimulus | ISO | J:164563 | |||||||||
| Biological Process | GO:0019221 | cytokine-mediated signaling pathway | IDA | J:7060 | |||||||||
| Biological Process | GO:0019221 | cytokine-mediated signaling pathway | IBA | J:265628 | |||||||||
| Biological Process | GO:0006897 | endocytosis | IEA | J:60000 | |||||||||
| Biological Process | GO:0060396 | growth hormone receptor signaling pathway | IDA | J:34907 | |||||||||
| Biological Process | GO:0060396 | growth hormone receptor signaling pathway | ISO | J:155856 | |||||||||
| Biological Process | GO:0060396 | growth hormone receptor signaling pathway | ISO | J:164563 | |||||||||
| Biological Process | GO:0060396 | growth hormone receptor signaling pathway | IBA | J:265628 | |||||||||
| Biological Process | GO:0042445 | hormone metabolic process | ISO | J:164563 | |||||||||
| Biological Process | GO:0009755 | hormone-mediated signaling pathway | ISO | J:155856 | |||||||||
| Biological Process | GO:0048009 | insulin-like growth factor receptor signaling pathway | ISO | J:164563 | |||||||||
| Biological Process | GO:0051179 | localization | ISO | J:164563 | |||||||||
| Biological Process | GO:1901215 | negative regulation of neuron death | ISO | J:155856 | |||||||||
| Biological Process | GO:0043406 | positive regulation of MAP kinase activity | ISO | J:155856 | |||||||||
| Biological Process | GO:0040018 | positive regulation of multicellular organism growth | ISO | J:164563 | |||||||||
| Biological Process | GO:0050731 | positive regulation of peptidyl-tyrosine phosphorylation | IDA | J:34907 | |||||||||
| Biological Process | GO:0050731 | positive regulation of peptidyl-tyrosine phosphorylation | IBA | J:265628 | |||||||||
| Biological Process | GO:0046427 | positive regulation of receptor signaling pathway via JAK-STAT | ISO | J:155856 | |||||||||
| Biological Process | GO:0046427 | positive regulation of receptor signaling pathway via JAK-STAT | IBA | J:265628 | |||||||||
| Biological Process | GO:0007259 | receptor signaling pathway via JAK-STAT | ISO | J:155856 | |||||||||
| Biological Process | GO:0040014 | regulation of multicellular organism growth | IMP | J:138238 | |||||||||
| Biological Process | GO:0032355 | response to estradiol | ISO | J:164563 | |||||||||
| Biological Process | GO:0019530 | taurine metabolic process | IMP | J:138238 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/23/2024 MGI 6.23 |

|
|
|
||