|
Symbol Name ID |
Gcnt1
glucosaminyl (N-acetyl) transferase 1, core 2 MGI:95676 |
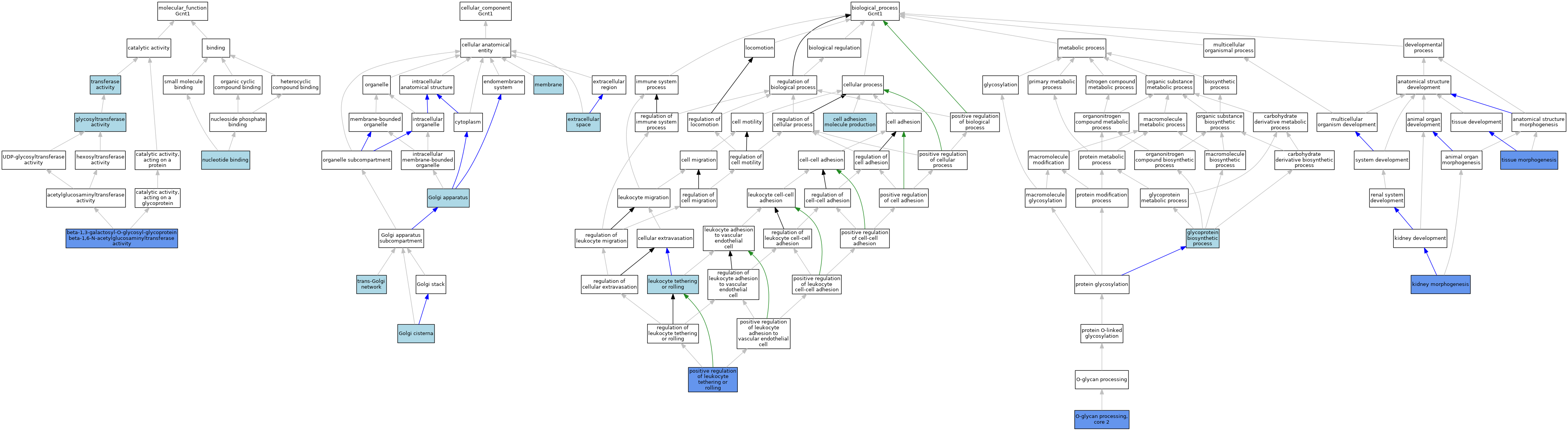
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity | IDA | J:320698 | |||||||||
| Molecular Function | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity | IDA | J:320156 | |||||||||
| Molecular Function | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity | ISO | J:155856 | |||||||||
| Molecular Function | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0016757 | glycosyltransferase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0016757 | glycosyltransferase activity | IEA | J:72247 | |||||||||
| Molecular Function | GO:0000166 | nucleotide binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0016740 | transferase activity | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005615 | extracellular space | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005794 | Golgi apparatus | IEA | J:60000 | |||||||||
| Cellular Component | GO:0031985 | Golgi cisterna | ISO | J:164563 | |||||||||
| Cellular Component | GO:0016020 | membrane | IEA | J:60000 | |||||||||
| Cellular Component | GO:0016020 | membrane | IEA | J:72247 | |||||||||
| Cellular Component | GO:0005802 | trans-Golgi network | ISO | J:164563 | |||||||||
| Biological Process | GO:0060352 | cell adhesion molecule production | ISO | J:164563 | |||||||||
| Biological Process | GO:0009101 | glycoprotein biosynthetic process | ISO | J:164563 | |||||||||
| Biological Process | GO:0060993 | kidney morphogenesis | IGI | J:149909 | |||||||||
| Biological Process | GO:0060993 | kidney morphogenesis | IGI | J:149909 | |||||||||
| Biological Process | GO:0050901 | leukocyte tethering or rolling | ISO | J:164563 | |||||||||
| Biological Process | GO:0016268 | O-glycan processing, core 2 | IMP | J:86387 | |||||||||
| Biological Process | GO:0016268 | O-glycan processing, core 2 | ISO | J:164563 | |||||||||
| Biological Process | GO:1903238 | positive regulation of leukocyte tethering or rolling | IMP | J:86387 | |||||||||
| Biological Process | GO:0048729 | tissue morphogenesis | IGI | J:149909 | |||||||||
| Biological Process | GO:0048729 | tissue morphogenesis | IGI | J:149909 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/16/2024 MGI 6.23 |

|
|
|
||