|
Symbol Name ID |
Flt3
FMS-like tyrosine kinase 3 MGI:95559 |
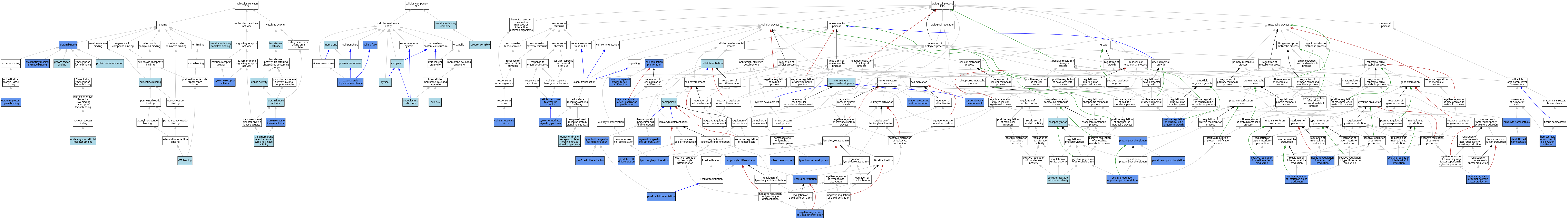
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0005524 | ATP binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0005524 | ATP binding | IEA | J:72247 | |||||||||
| Molecular Function | GO:0004896 | cytokine receptor activity | IMP | J:136209 | |||||||||
| Molecular Function | GO:0019838 | growth factor binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:0016301 | kinase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0035259 | nuclear glucocorticoid receptor binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0000166 | nucleotide binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0043548 | phosphatidylinositol 3-kinase binding | IDA | J:18780 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:56367 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:18780 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:36245 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:162445 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:16458 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:23709 | |||||||||
| Molecular Function | GO:0004672 | protein kinase activity | IEA | J:72247 | |||||||||
| Molecular Function | GO:0043621 | protein self-association | ISO | J:164563 | |||||||||
| Molecular Function | GO:0004713 | protein tyrosine kinase activity | IDA | J:140548 | |||||||||
| Molecular Function | GO:0044877 | protein-containing complex binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0016740 | transferase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0004714 | transmembrane receptor protein tyrosine kinase activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0031625 | ubiquitin protein ligase binding | IPI | J:213100 | |||||||||
| Cellular Component | GO:0009986 | cell surface | IDA | J:47896 | |||||||||
| Cellular Component | GO:0009986 | cell surface | IDA | J:27743 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005829 | cytosol | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005783 | endoplasmic reticulum | ISO | J:164563 | |||||||||
| Cellular Component | GO:0009897 | external side of plasma membrane | IDA | J:203580 | |||||||||
| Cellular Component | GO:0016020 | membrane | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005634 | nucleus | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | TAS | Reactome:R-MMU-9604768 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | TAS | Reactome:R-MMU-9604980 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | TAS | Reactome:R-MMU-9605258 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | TAS | Reactome:R-MMU-9606621 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | TAS | Reactome:R-MMU-9606623 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | TAS | Reactome:R-MMU-9606789 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | TAS | Reactome:R-MMU-9607226 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | TAS | Reactome:R-MMU-9609272 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | TAS | Reactome:R-MMU-9645128 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | TAS | Reactome:R-MMU-9645133 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | TAS | Reactome:R-MMU-9645136 | |||||||||
| Cellular Component | GO:0032991 | protein-containing complex | ISO | J:155856 | |||||||||
| Cellular Component | GO:0043235 | receptor complex | IBA | J:265628 | |||||||||
| Biological Process | GO:0019882 | antigen processing and presentation | IMP | J:159599 | |||||||||
| Biological Process | GO:0030183 | B cell differentiation | IMP | J:170520 | |||||||||
| Biological Process | GO:0030183 | B cell differentiation | IBA | J:265628 | |||||||||
| Biological Process | GO:0030154 | cell differentiation | IEA | J:72247 | |||||||||
| Biological Process | GO:0008283 | cell population proliferation | IMP | J:21273 | |||||||||
| Biological Process | GO:0071345 | cellular response to cytokine stimulus | IMP | J:136209 | |||||||||
| Biological Process | GO:0098586 | cellular response to virus | IMP | J:159599 | |||||||||
| Biological Process | GO:0035726 | common myeloid progenitor cell proliferation | IMP | J:177063 | |||||||||
| Biological Process | GO:0019221 | cytokine-mediated signaling pathway | IMP | J:136209 | |||||||||
| Biological Process | GO:0019221 | cytokine-mediated signaling pathway | IBA | J:265628 | |||||||||
| Biological Process | GO:0097028 | dendritic cell differentiation | IMP | J:136209 | |||||||||
| Biological Process | GO:0036145 | dendritic cell homeostasis | IMP | J:159599 | |||||||||
| Biological Process | GO:0030097 | hemopoiesis | ISO | J:16485 | |||||||||
| Biological Process | GO:0048873 | homeostasis of number of cells within a tissue | IGI | J:189096 | |||||||||
| Biological Process | GO:0048873 | homeostasis of number of cells within a tissue | IMP | J:159599 | |||||||||
| Biological Process | GO:0001776 | leukocyte homeostasis | IMP | J:136209 | |||||||||
| Biological Process | GO:0048535 | lymph node development | IMP | J:159599 | |||||||||
| Biological Process | GO:0030098 | lymphocyte differentiation | IDA | J:79569 | |||||||||
| Biological Process | GO:0046651 | lymphocyte proliferation | IMP | J:136209 | |||||||||
| Biological Process | GO:0002320 | lymphoid progenitor cell differentiation | IGI | J:27238 | |||||||||
| Biological Process | GO:0007275 | multicellular organism development | IBA | J:265628 | |||||||||
| Biological Process | GO:0002318 | myeloid progenitor cell differentiation | IMP | J:136209 | |||||||||
| Biological Process | GO:0002318 | myeloid progenitor cell differentiation | IMP | J:133528 | |||||||||
| Biological Process | GO:0002318 | myeloid progenitor cell differentiation | IGI | J:27238 | |||||||||
| Biological Process | GO:0045578 | negative regulation of B cell differentiation | IDA | J:108330 | |||||||||
| Biological Process | GO:0008285 | negative regulation of cell population proliferation | IMP | J:21273 | |||||||||
| Biological Process | GO:0032715 | negative regulation of interleukin-6 production | IMP | J:159599 | |||||||||
| Biological Process | GO:0032720 | negative regulation of tumor necrosis factor production | IMP | J:159599 | |||||||||
| Biological Process | GO:0016310 | phosphorylation | IEA | J:60000 | |||||||||
| Biological Process | GO:0032727 | positive regulation of interferon-alpha production | IMP | J:159599 | |||||||||
| Biological Process | GO:0032735 | positive regulation of interleukin-12 production | IMP | J:159599 | |||||||||
| Biological Process | GO:0033674 | positive regulation of kinase activity | IBA | J:265628 | |||||||||
| Biological Process | GO:0040018 | positive regulation of multicellular organism growth | IMP | J:159599 | |||||||||
| Biological Process | GO:0001934 | positive regulation of protein phosphorylation | IGI | J:162445 | |||||||||
| Biological Process | GO:0001934 | positive regulation of protein phosphorylation | IGI | J:162445 | |||||||||
| Biological Process | GO:0032729 | positive regulation of type II interferon production | IMP | J:159599 | |||||||||
| Biological Process | GO:0009791 | post-embryonic development | IGI | J:189096 | |||||||||
| Biological Process | GO:0002328 | pro-B cell differentiation | IMP | J:170520 | |||||||||
| Biological Process | GO:0002328 | pro-B cell differentiation | IMP | J:27238 | |||||||||
| Biological Process | GO:0002572 | pro-T cell differentiation | IMP | J:27238 | |||||||||
| Biological Process | GO:0046777 | protein autophosphorylation | IDA | J:18780 | |||||||||
| Biological Process | GO:0046777 | protein autophosphorylation | IDA | J:140548 | |||||||||
| Biological Process | GO:0006468 | protein phosphorylation | IGI | J:162445 | |||||||||
| Biological Process | GO:0006468 | protein phosphorylation | IGI | J:162445 | |||||||||
| Biological Process | GO:0048536 | spleen development | IGI | J:189096 | |||||||||
| Biological Process | GO:0007169 | transmembrane receptor protein tyrosine kinase signaling pathway | IBA | J:265628 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/30/2024 MGI 6.23 |

|
|
|
||