|
Symbol Name ID |
Dpp4
dipeptidylpeptidase 4 MGI:94919 |
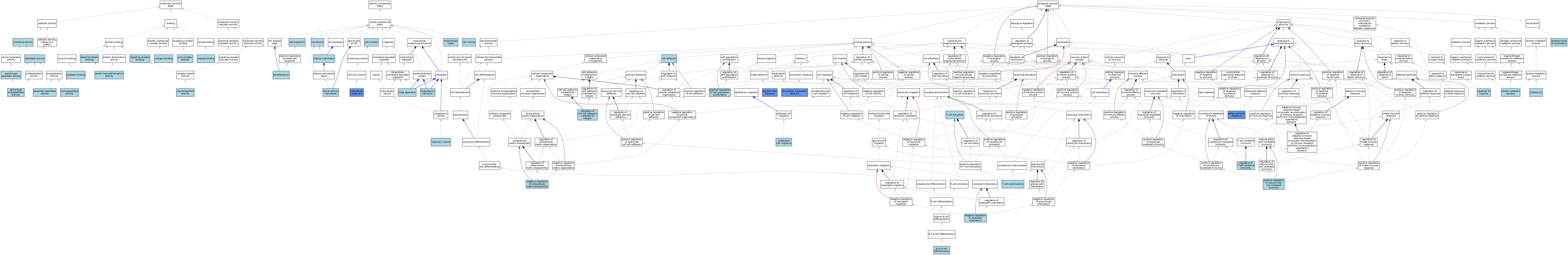
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0004177 | aminopeptidase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0045499 | chemorepellent activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0005518 | collagen binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0008239 | dipeptidyl-peptidase activity | ISO | J:155856 | |||||||||
| Molecular Function | GO:0008239 | dipeptidyl-peptidase activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0008239 | dipeptidyl-peptidase activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0016787 | hydrolase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0042802 | identical protein binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0042802 | identical protein binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0008233 | peptidase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0042277 | peptide binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0002020 | protease binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0042803 | protein homodimerization activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0004252 | serine-type endopeptidase activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0008236 | serine-type peptidase activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0005102 | signaling receptor binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0001618 | virus receptor activity | ISO | J:164563 | |||||||||
| Cellular Component | GO:0016324 | apical plasma membrane | ISO | J:164563 | |||||||||
| Cellular Component | GO:0030054 | cell junction | IEA | J:60000 | |||||||||
| Cellular Component | GO:0042995 | cell projection | IEA | J:60000 | |||||||||
| Cellular Component | GO:0009986 | cell surface | ISO | J:164563 | |||||||||
| Cellular Component | GO:0009986 | cell surface | ISO | J:155856 | |||||||||
| Cellular Component | GO:0030139 | endocytic vesicle | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005783 | endoplasmic reticulum | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005576 | extracellular region | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005794 | Golgi apparatus | ISO | J:155856 | |||||||||
| Cellular Component | GO:0046581 | intercellular canaliculus | IDA | J:77485 | |||||||||
| Cellular Component | GO:0030027 | lamellipodium | ISO | J:164563 | |||||||||
| Cellular Component | GO:0016020 | membrane | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | IBA | J:265628 | |||||||||
| Biological Process | GO:0002337 | B-1a B cell differentiation | ISO | J:155856 | |||||||||
| Biological Process | GO:0001662 | behavioral fear response | IMP | J:110634 | |||||||||
| Biological Process | GO:0007155 | cell adhesion | IEA | J:60000 | |||||||||
| Biological Process | GO:0043542 | endothelial cell migration | ISO | J:164563 | |||||||||
| Biological Process | GO:0051234 | establishment of localization | ISO | J:155856 | |||||||||
| Biological Process | GO:0035641 | locomotory exploration behavior | IMP | J:110634 | |||||||||
| Biological Process | GO:0010716 | negative regulation of extracellular matrix disassembly | ISO | J:164563 | |||||||||
| Biological Process | GO:0090024 | negative regulation of neutrophil chemotaxis | ISO | J:164563 | |||||||||
| Biological Process | GO:0008284 | positive regulation of cell population proliferation | ISO | J:164563 | |||||||||
| Biological Process | GO:0002717 | positive regulation of natural killer cell mediated immunity | ISO | J:155856 | |||||||||
| Biological Process | GO:0030163 | protein catabolic process | ISO | J:155856 | |||||||||
| Biological Process | GO:0006508 | proteolysis | ISO | J:164563 | |||||||||
| Biological Process | GO:0006508 | proteolysis | IBA | J:265628 | |||||||||
| Biological Process | GO:0036343 | psychomotor behavior | IMP | J:110634 | |||||||||
| Biological Process | GO:0033632 | regulation of cell-cell adhesion mediated by integrin | ISO | J:164563 | |||||||||
| Biological Process | GO:0002709 | regulation of T cell mediated immunity | ISO | J:155856 | |||||||||
| Biological Process | GO:0001666 | response to hypoxia | ISO | J:164563 | |||||||||
| Biological Process | GO:0042110 | T cell activation | ISO | J:164563 | |||||||||
| Biological Process | GO:0031295 | T cell costimulation | ISO | J:164563 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/16/2024 MGI 6.23 |

|
|
|
||