|
Symbol Name ID |
Cd247
CD247 antigen MGI:88334 |
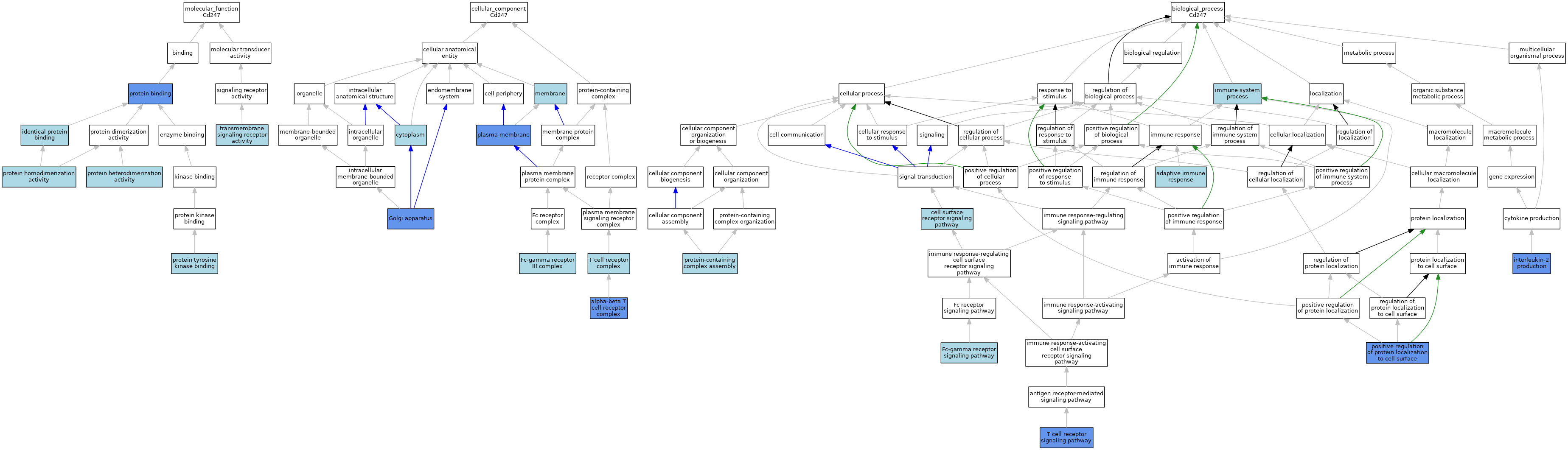
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0042802 | identical protein binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:209882 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:137841 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:138871 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:204615 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:18500 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:91455 | |||||||||
| Molecular Function | GO:0046982 | protein heterodimerization activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0042803 | protein homodimerization activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:1990782 | protein tyrosine kinase binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0004888 | transmembrane signaling receptor activity | IEA | J:72247 | |||||||||
| Cellular Component | GO:0042105 | alpha-beta T cell receptor complex | ISO | J:164563 | |||||||||
| Cellular Component | GO:0042105 | alpha-beta T cell receptor complex | IBA | J:265628 | |||||||||
| Cellular Component | GO:0042105 | alpha-beta T cell receptor complex | IDA | J:35279 | |||||||||
| Cellular Component | GO:0042105 | alpha-beta T cell receptor complex | IDA | J:41170 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | ISO | J:73065 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | ISO | J:91455 | |||||||||
| Cellular Component | GO:0033001 | Fc-gamma receptor III complex | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005794 | Golgi apparatus | IDA | J:297527 | |||||||||
| Cellular Component | GO:0016020 | membrane | IEA | J:60000 | |||||||||
| Cellular Component | GO:0016020 | membrane | IEA | J:72247 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | IDA | J:297527 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | ISO | J:73065 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | ISO | J:91455 | |||||||||
| Cellular Component | GO:0042101 | T cell receptor complex | ISO | J:155856 | |||||||||
| Cellular Component | GO:0042101 | T cell receptor complex | ISO | J:73065 | |||||||||
| Biological Process | GO:0002250 | adaptive immune response | IEA | J:60000 | |||||||||
| Biological Process | GO:0007166 | cell surface receptor signaling pathway | ISO | J:155856 | |||||||||
| Biological Process | GO:0038094 | Fc-gamma receptor signaling pathway | ISO | J:164563 | |||||||||
| Biological Process | GO:0002376 | immune system process | IEA | J:60000 | |||||||||
| Biological Process | GO:0032623 | interleukin-2 production | IGI | J:113761 | |||||||||
| Biological Process | GO:2000010 | positive regulation of protein localization to cell surface | IDA | J:113761 | |||||||||
| Biological Process | GO:0065003 | protein-containing complex assembly | ISO | J:164563 | |||||||||
| Biological Process | GO:0050852 | T cell receptor signaling pathway | IBA | J:265628 | |||||||||
| Biological Process | GO:0050852 | T cell receptor signaling pathway | IDA | J:110739 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/16/2024 MGI 6.23 |

|
|
|
||