|
Symbol Name ID |
Alox15
arachidonate 15-lipoxygenase MGI:87997 |
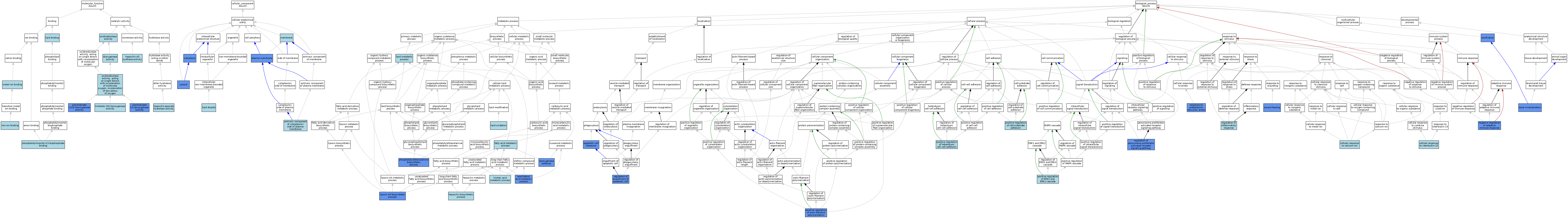
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0004052 | arachidonate 12(S)-lipoxygenase activity | ISO | J:155856 | |||||||||
| Molecular Function | GO:0004052 | arachidonate 12(S)-lipoxygenase activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0004052 | arachidonate 12(S)-lipoxygenase activity | IDA | J:18190 | |||||||||
| Molecular Function | GO:0004052 | arachidonate 12(S)-lipoxygenase activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0050473 | arachidonate 15-lipoxygenase activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0050473 | arachidonate 15-lipoxygenase activity | IDA | J:18190 | |||||||||
| Molecular Function | GO:0050473 | arachidonate 15-lipoxygenase activity | ISO | J:155856 | |||||||||
| Molecular Function | GO:0050473 | arachidonate 15-lipoxygenase activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0051213 | dioxygenase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0051120 | hepoxilin A3 synthase activity | ISO | J:155856 | |||||||||
| Molecular Function | GO:0047977 | hepoxilin-epoxide hydrolase activity | ISO | J:155856 | |||||||||
| Molecular Function | GO:0005506 | iron ion binding | IEA | J:72247 | |||||||||
| Molecular Function | GO:0016165 | linoleate 13S-lipoxygenase activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0008289 | lipid binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0046872 | metal ion binding | IEA | J:72247 | |||||||||
| Molecular Function | GO:0046872 | metal ion binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0016491 | oxidoreductase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen | IEA | J:72247 | |||||||||
| Molecular Function | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IDA | J:120937 | |||||||||
| Cellular Component | GO:0005829 | cytosol | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005829 | cytosol | IDA | J:200335 | |||||||||
| Cellular Component | GO:0005829 | cytosol | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005829 | cytosol | IBA | J:265628 | |||||||||
| Cellular Component | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005811 | lipid droplet | ISO | J:164563 | |||||||||
| Cellular Component | GO:0016020 | membrane | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | IDA | J:200335 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | IBA | J:265628 | |||||||||
| Biological Process | GO:0043277 | apoptotic cell clearance | IMP | J:187327 | |||||||||
| Biological Process | GO:0019369 | arachidonic acid metabolic process | IMP | J:44778 | |||||||||
| Biological Process | GO:0019369 | arachidonic acid metabolic process | ISO | J:155856 | |||||||||
| Biological Process | GO:0019369 | arachidonic acid metabolic process | IDA | J:18190 | |||||||||
| Biological Process | GO:0019369 | arachidonic acid metabolic process | ISO | J:164563 | |||||||||
| Biological Process | GO:0030282 | bone mineralization | IMP | J:87403 | |||||||||
| Biological Process | GO:0071277 | cellular response to calcium ion | ISO | J:164563 | |||||||||
| Biological Process | GO:0035963 | cellular response to interleukin-13 | ISO | J:164563 | |||||||||
| Biological Process | GO:0006631 | fatty acid metabolic process | IEA | J:60000 | |||||||||
| Biological Process | GO:0051122 | hepoxilin biosynthetic process | ISO | J:155856 | |||||||||
| Biological Process | GO:0043651 | linoleic acid metabolic process | ISO | J:164563 | |||||||||
| Biological Process | GO:0043651 | linoleic acid metabolic process | IBA | J:265628 | |||||||||
| Biological Process | GO:0006629 | lipid metabolic process | ISO | J:164563 | |||||||||
| Biological Process | GO:0034440 | lipid oxidation | IBA | J:265628 | |||||||||
| Biological Process | GO:2001303 | lipoxin A4 biosynthetic process | IMP | J:98733 | |||||||||
| Biological Process | GO:0019372 | lipoxygenase pathway | ISO | J:164563 | |||||||||
| Biological Process | GO:0019372 | lipoxygenase pathway | ISO | J:155856 | |||||||||
| Biological Process | GO:0019372 | lipoxygenase pathway | IDA | J:18190 | |||||||||
| Biological Process | GO:0019372 | lipoxygenase pathway | IMP | J:44778 | |||||||||
| Biological Process | GO:0002820 | negative regulation of adaptive immune response | IMP | J:187327 | |||||||||
| Biological Process | GO:0001503 | ossification | IMP | J:87403 | |||||||||
| Biological Process | GO:0006646 | phosphatidylethanolamine biosynthetic process | IMP | J:187327 | |||||||||
| Biological Process | GO:0030838 | positive regulation of actin filament polymerization | IMP | J:200335 | |||||||||
| Biological Process | GO:0010811 | positive regulation of cell-substrate adhesion | ISO | J:164563 | |||||||||
| Biological Process | GO:0070374 | positive regulation of ERK1 and ERK2 cascade | ISO | J:164563 | |||||||||
| Biological Process | GO:0034116 | positive regulation of heterotypic cell-cell adhesion | ISO | J:155856 | |||||||||
| Biological Process | GO:1901074 | regulation of engulfment of apoptotic cell | IMP | J:187327 | |||||||||
| Biological Process | GO:1901074 | regulation of engulfment of apoptotic cell | IMP | J:200335 | |||||||||
| Biological Process | GO:0050727 | regulation of inflammatory response | ISO | J:164563 | |||||||||
| Biological Process | GO:0035358 | regulation of peroxisome proliferator activated receptor signaling pathway | IMP | J:200339 | |||||||||
| Biological Process | GO:0034976 | response to endoplasmic reticulum stress | IMP | J:184646 | |||||||||
| Biological Process | GO:0042060 | wound healing | IMP | J:98733 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/30/2024 MGI 6.23 |

|
|
|
||