|
Symbol Name ID |
Ak2
adenylate kinase 2 MGI:87978 |
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
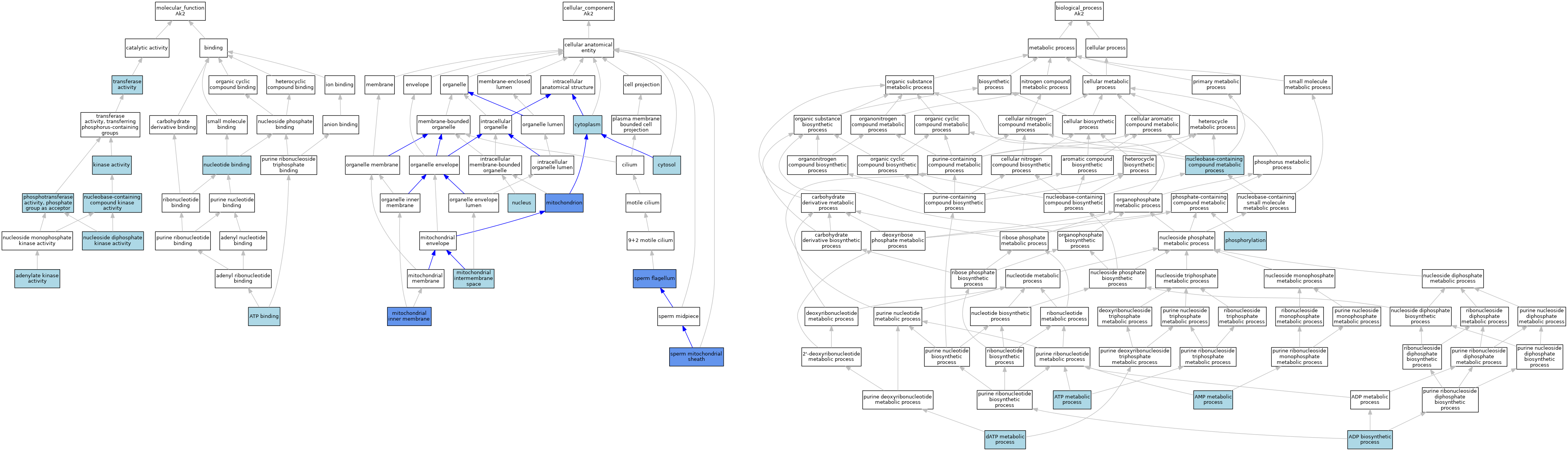
| Molecular Function | GO:0004017 | adenylate kinase activity | ISO | J:155856 | |||||||||
| Molecular Function | GO:0004017 | adenylate kinase activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0004017 | adenylate kinase activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0005524 | ATP binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0005524 | ATP binding | IEA | J:72247 | |||||||||
| Molecular Function | GO:0016301 | kinase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0019205 | nucleobase-containing compound kinase activity | ISO | J:155856 | |||||||||
| Molecular Function | GO:0004550 | nucleoside diphosphate kinase activity | IEA | J:72245 | |||||||||
| Molecular Function | GO:0000166 | nucleotide binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0016776 | phosphotransferase activity, phosphate group as acceptor | IEA | J:72247 | |||||||||
| Molecular Function | GO:0016740 | transferase activity | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005829 | cytosol | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005743 | mitochondrial inner membrane | HDA | J:100953 | |||||||||
| Cellular Component | GO:0005758 | mitochondrial intermembrane space | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005739 | mitochondrion | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005739 | mitochondrion | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005739 | mitochondrion | HDA | J:151002 | |||||||||
| Cellular Component | GO:0005739 | mitochondrion | HDA | J:86816 | |||||||||
| Cellular Component | GO:0005739 | mitochondrion | HDA | J:86816 | |||||||||
| Cellular Component | GO:0005739 | mitochondrion | HDA | J:86816 | |||||||||
| Cellular Component | GO:0005739 | mitochondrion | HDA | J:86816 | |||||||||
| Cellular Component | GO:0005634 | nucleus | ISO | J:155856 | |||||||||
| Cellular Component | GO:0036126 | sperm flagellum | IDA | J:113416 | |||||||||
| Cellular Component | GO:0097226 | sperm mitochondrial sheath | IDA | J:113416 | |||||||||
| Biological Process | GO:0006172 | ADP biosynthetic process | ISO | J:155856 | |||||||||
| Biological Process | GO:0046033 | AMP metabolic process | ISO | J:155856 | |||||||||
| Biological Process | GO:0046034 | ATP metabolic process | ISO | J:155856 | |||||||||
| Biological Process | GO:0046060 | dATP metabolic process | ISO | J:155856 | |||||||||
| Biological Process | GO:0006139 | nucleobase-containing compound metabolic process | IEA | J:72247 | |||||||||
| Biological Process | GO:0016310 | phosphorylation | IEA | J:60000 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/16/2024 MGI 6.23 |

|
|
|
||