|
Symbol Name ID |
Adh5
alcohol dehydrogenase 5 (class III), chi polypeptide MGI:87929 |
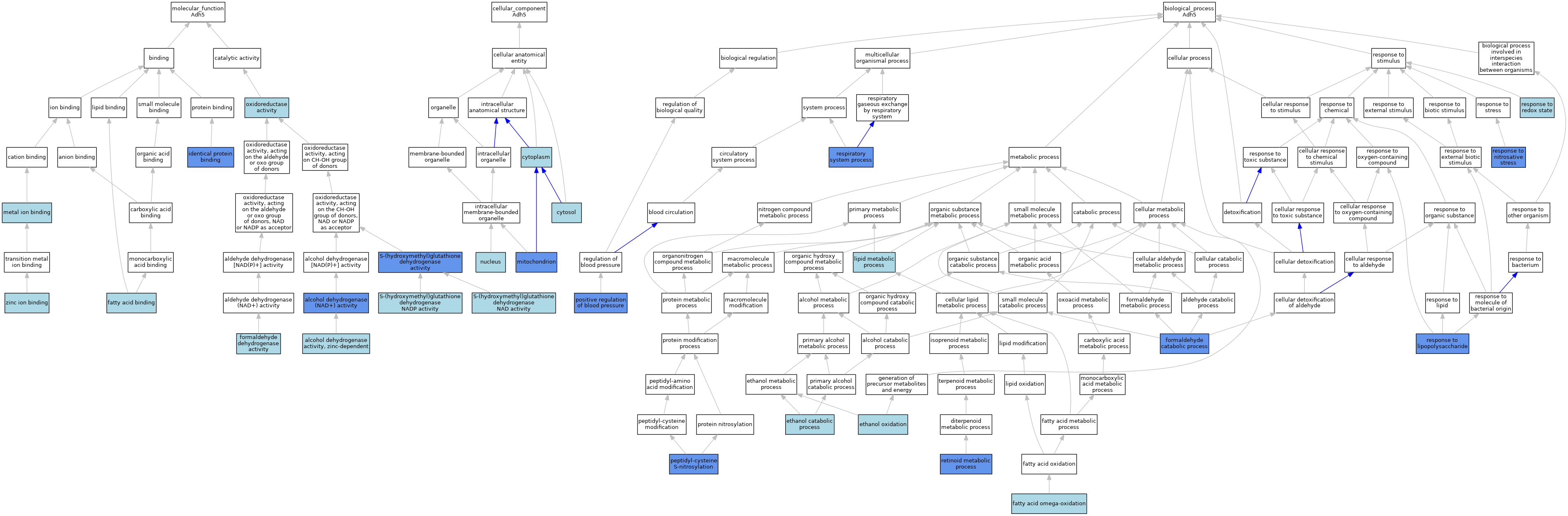
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0004022 | alcohol dehydrogenase (NAD+) activity | ISO | J:155856 | |||||||||
| Molecular Function | GO:0004022 | alcohol dehydrogenase (NAD+) activity | IDA | J:7858 | |||||||||
| Molecular Function | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent | IBA | J:265628 | |||||||||
| Molecular Function | GO:0005504 | fatty acid binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0018467 | formaldehyde dehydrogenase activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0042802 | identical protein binding | IPI | J:7858 | |||||||||
| Molecular Function | GO:0046872 | metal ion binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0016491 | oxidoreductase activity | IEA | J:72247 | |||||||||
| Molecular Function | GO:0016491 | oxidoreductase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0051903 | S-(hydroxymethyl)glutathione dehydrogenase activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0051903 | S-(hydroxymethyl)glutathione dehydrogenase activity | IMP | J:105651 | |||||||||
| Molecular Function | GO:0106322 | S-(hydroxymethyl)glutathione dehydrogenase NAD activity | IEA | J:72245 | |||||||||
| Molecular Function | GO:0106321 | S-(hydroxymethyl)glutathione dehydrogenase NADP activity | IEA | J:72245 | |||||||||
| Molecular Function | GO:0008270 | zinc ion binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0008270 | zinc ion binding | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005829 | cytosol | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005739 | mitochondrion | IDA | J:7858 | |||||||||
| Cellular Component | GO:0005634 | nucleus | ISO | J:155856 | |||||||||
| Biological Process | GO:0006068 | ethanol catabolic process | ISO | J:155856 | |||||||||
| Biological Process | GO:0006069 | ethanol oxidation | IEA | J:72247 | |||||||||
| Biological Process | GO:0010430 | fatty acid omega-oxidation | ISO | J:164563 | |||||||||
| Biological Process | GO:0046294 | formaldehyde catabolic process | IBA | J:265628 | |||||||||
| Biological Process | GO:0046294 | formaldehyde catabolic process | IMP | J:55555 | |||||||||
| Biological Process | GO:0006629 | lipid metabolic process | IEA | J:60000 | |||||||||
| Biological Process | GO:0018119 | peptidyl-cysteine S-nitrosylation | IMP | J:88337 | |||||||||
| Biological Process | GO:0045777 | positive regulation of blood pressure | IMP | J:88337 | |||||||||
| Biological Process | GO:0003016 | respiratory system process | IMP | J:105651 | |||||||||
| Biological Process | GO:0032496 | response to lipopolysaccharide | IMP | J:88337 | |||||||||
| Biological Process | GO:0051409 | response to nitrosative stress | IMP | J:88337 | |||||||||
| Biological Process | GO:0051775 | response to redox state | ISO | J:164563 | |||||||||
| Biological Process | GO:0001523 | retinoid metabolic process | IMP | J:76746 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/16/2024 MGI 6.23 |

|
|
|
||