|
Symbol Name ID |
Il1rapl1
interleukin 1 receptor accessory protein-like 1 MGI:2687319 |
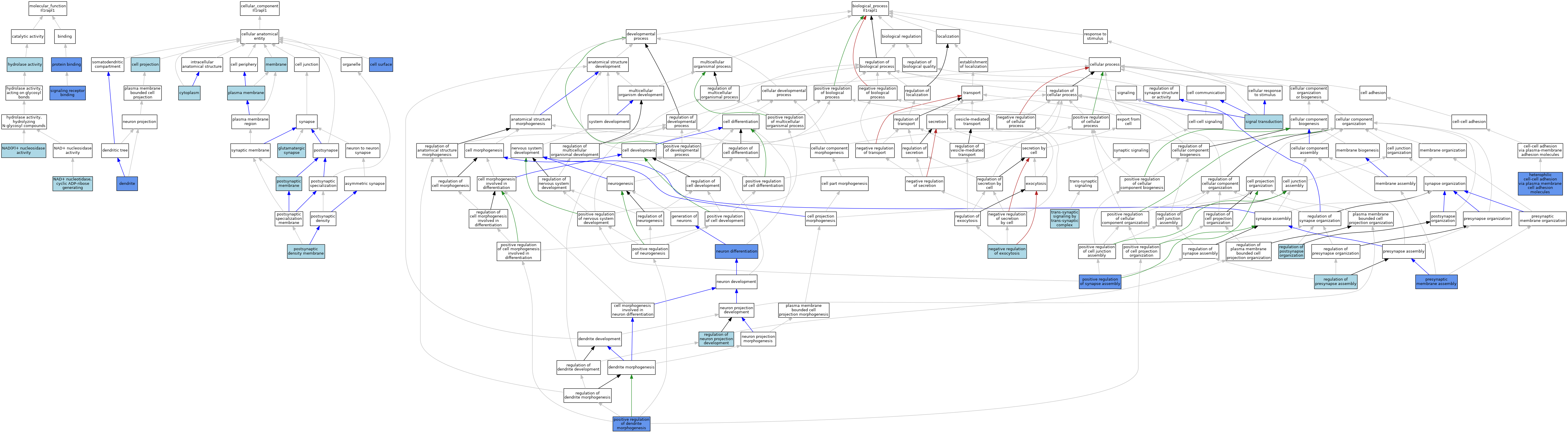
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0016787 | hydrolase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0050135 | NAD(P)+ nucleosidase activity | IEA | J:72245 | |||||||||
| Molecular Function | GO:0061809 | NAD+ nucleotidase, cyclic ADP-ribose generating | IEA | J:72245 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:177124 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:222796 | |||||||||
| Molecular Function | GO:0005102 | signaling receptor binding | IPI | J:177124 | |||||||||
| Cellular Component | GO:0042995 | cell projection | IEA | J:60000 | |||||||||
| Cellular Component | GO:0009986 | cell surface | IDA | J:177124 | |||||||||
| Cellular Component | GO:0009986 | cell surface | IDA | J:177124 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IEA | J:60000 | |||||||||
| Cellular Component | GO:0030425 | dendrite | IDA | J:177124 | |||||||||
| Cellular Component | GO:0030425 | dendrite | IDA | J:177124 | |||||||||
| Cellular Component | GO:0098978 | glutamatergic synapse | ISO | J:164563 | |||||||||
| Cellular Component | GO:0098978 | glutamatergic synapse | ISO | J:155856 | |||||||||
| Cellular Component | GO:0016020 | membrane | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | ISO | J:164563 | |||||||||
| Cellular Component | GO:0098839 | postsynaptic density membrane | ISO | J:155856 | |||||||||
| Cellular Component | GO:0045211 | postsynaptic membrane | NAS | J:177124 | |||||||||
| Biological Process | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules | IDA | J:177124 | |||||||||
| Biological Process | GO:0045920 | negative regulation of exocytosis | ISO | J:155856 | |||||||||
| Biological Process | GO:0045920 | negative regulation of exocytosis | ISO | J:164563 | |||||||||
| Biological Process | GO:0045920 | negative regulation of exocytosis | IBA | J:265628 | |||||||||
| Biological Process | GO:0030182 | neuron differentiation | IDA | J:177124 | |||||||||
| Biological Process | GO:0050775 | positive regulation of dendrite morphogenesis | IMP | J:177124 | |||||||||
| Biological Process | GO:0051965 | positive regulation of synapse assembly | IDA | J:222796 | |||||||||
| Biological Process | GO:0097105 | presynaptic membrane assembly | IMP | J:177124 | |||||||||
| Biological Process | GO:0010975 | regulation of neuron projection development | ISO | J:155856 | |||||||||
| Biological Process | GO:0099175 | regulation of postsynapse organization | ISO | J:164563 | |||||||||
| Biological Process | GO:1905606 | regulation of presynapse assembly | ISO | J:164563 | |||||||||
| Biological Process | GO:0007165 | signal transduction | IEA | J:72247 | |||||||||
| Biological Process | GO:0099545 | trans-synaptic signaling by trans-synaptic complex | ISO | J:164563 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/16/2024 MGI 6.23 |

|
|
|
||