|
Symbol Name ID |
Rc3h1
RING CCCH (C3H) domains 1 MGI:2685397 |
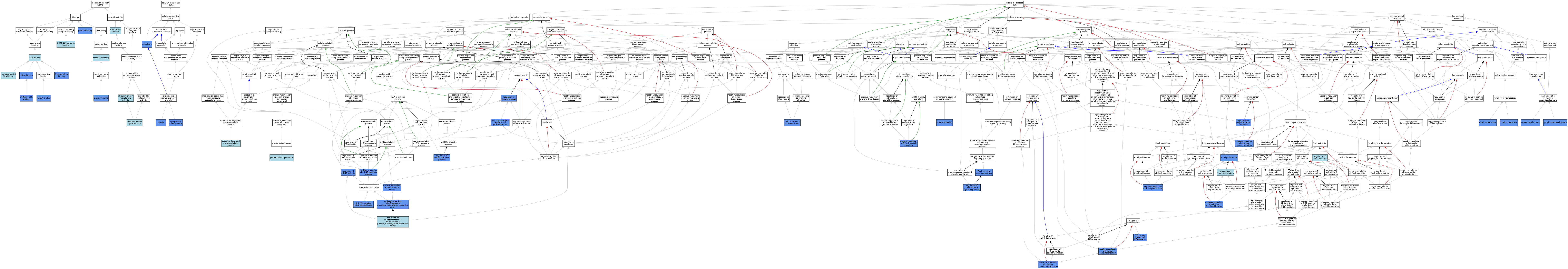
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:1905762 | CCR4-NOT complex binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0003725 | double-stranded RNA binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0046872 | metal ion binding | IEA | J:72247 | |||||||||
| Molecular Function | GO:0046872 | metal ion binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0035198 | miRNA binding | IDA | J:221791 | |||||||||
| Molecular Function | GO:0003730 | mRNA 3'-UTR binding | IDA | J:229326 | |||||||||
| Molecular Function | GO:0003730 | mRNA 3'-UTR binding | IDA | J:162534 | |||||||||
| Molecular Function | GO:0003730 | mRNA 3'-UTR binding | IDA | J:228359 | |||||||||
| Molecular Function | GO:0003729 | mRNA binding | IDA | J:196136 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:162534 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:221791 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:196136 | |||||||||
| Molecular Function | GO:0003723 | RNA binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0035613 | RNA stem-loop binding | IDA | J:261058 | |||||||||
| Molecular Function | GO:0035613 | RNA stem-loop binding | IDA | J:228359 | |||||||||
| Molecular Function | GO:0035613 | RNA stem-loop binding | IDA | J:229326 | |||||||||
| Molecular Function | GO:0035613 | RNA stem-loop binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0016740 | transferase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0061630 | ubiquitin protein ligase activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0004842 | ubiquitin-protein transferase activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0008270 | zinc ion binding | IDA | J:221791 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IDA | J:98938 | |||||||||
| Cellular Component | GO:0010494 | cytoplasmic stress granule | IDA | J:228359 | |||||||||
| Cellular Component | GO:0010494 | cytoplasmic stress granule | IDA | J:98938 | |||||||||
| Cellular Component | GO:0010494 | cytoplasmic stress granule | IDA | J:162534 | |||||||||
| Cellular Component | GO:0010494 | cytoplasmic stress granule | IDA | J:229326 | |||||||||
| Cellular Component | GO:0000932 | P-body | IDA | J:229326 | |||||||||
| Cellular Component | GO:0000932 | P-body | IDA | J:162534 | |||||||||
| Cellular Component | GO:0000932 | P-body | ISO | J:164563 | |||||||||
| Cellular Component | GO:0000932 | P-body | IDA | J:228359 | |||||||||
| Biological Process | GO:0061158 | 3'-UTR-mediated mRNA destabilization | IDA | J:229326 | |||||||||
| Biological Process | GO:0061158 | 3'-UTR-mediated mRNA destabilization | IDA | J:228359 | |||||||||
| Biological Process | GO:0001782 | B cell homeostasis | IGI | J:196136 | |||||||||
| Biological Process | GO:0071347 | cellular response to interleukin-1 | IDA | J:229326 | |||||||||
| Biological Process | GO:0048535 | lymph node development | IGI | J:196136 | |||||||||
| Biological Process | GO:0046007 | negative regulation of activated T cell proliferation | IMP | J:175161 | |||||||||
| Biological Process | GO:0030889 | negative regulation of B cell proliferation | IMP | J:175161 | |||||||||
| Biological Process | GO:0002635 | negative regulation of germinal center formation | IMP | J:175161 | |||||||||
| Biological Process | GO:2000320 | negative regulation of T-helper 17 cell differentiation | IMP | J:259145 | |||||||||
| Biological Process | GO:0045623 | negative regulation of T-helper cell differentiation | IMP | J:98938 | |||||||||
| Biological Process | GO:0000956 | nuclear-transcribed mRNA catabolic process | IMP | J:162534 | |||||||||
| Biological Process | GO:0000956 | nuclear-transcribed mRNA catabolic process | ISO | J:164563 | |||||||||
| Biological Process | GO:0000288 | nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay | IMP | J:229326 | |||||||||
| Biological Process | GO:0000288 | nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay | IMP | J:228359 | |||||||||
| Biological Process | GO:0000288 | nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay | IBA | J:265628 | |||||||||
| Biological Process | GO:0033962 | P-body assembly | IMP | J:162534 | |||||||||
| Biological Process | GO:0061014 | positive regulation of mRNA catabolic process | IDA | J:229326 | |||||||||
| Biological Process | GO:0061014 | positive regulation of mRNA catabolic process | IDA | J:228359 | |||||||||
| Biological Process | GO:0061014 | positive regulation of mRNA catabolic process | IMP | J:261058 | |||||||||
| Biological Process | GO:1901224 | positive regulation of NIK/NF-kappaB signaling | IDA | J:196136 | |||||||||
| Biological Process | GO:0010608 | post-transcriptional regulation of gene expression | IMP | J:261058 | |||||||||
| Biological Process | GO:0010608 | post-transcriptional regulation of gene expression | IMP | J:162534 | |||||||||
| Biological Process | GO:0010608 | post-transcriptional regulation of gene expression | IDA | J:196136 | |||||||||
| Biological Process | GO:0000209 | protein polyubiquitination | ISO | J:164563 | |||||||||
| Biological Process | GO:0010468 | regulation of gene expression | IDA | J:196136 | |||||||||
| Biological Process | GO:0002634 | regulation of germinal center formation | IMP | J:98938 | |||||||||
| Biological Process | GO:0002634 | regulation of germinal center formation | IMP | J:146622 | |||||||||
| Biological Process | GO:2000628 | regulation of miRNA metabolic process | IMP | J:221791 | |||||||||
| Biological Process | GO:2000628 | regulation of miRNA metabolic process | IMP | J:281595 | |||||||||
| Biological Process | GO:0043488 | regulation of mRNA stability | IMP | J:175161 | |||||||||
| Biological Process | GO:1900151 | regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay | ISO | J:164563 | |||||||||
| Biological Process | GO:0050863 | regulation of T cell activation | IEA | J:72247 | |||||||||
| Biological Process | GO:0042129 | regulation of T cell proliferation | IEA | J:72247 | |||||||||
| Biological Process | GO:0050856 | regulation of T cell receptor signaling pathway | IMP | J:98938 | |||||||||
| Biological Process | GO:0048536 | spleen development | IGI | J:196136 | |||||||||
| Biological Process | GO:0043029 | T cell homeostasis | IGI | J:196136 | |||||||||
| Biological Process | GO:0042098 | T cell proliferation | IGI | J:196136 | |||||||||
| Biological Process | GO:0050852 | T cell receptor signaling pathway | IMP | J:259145 | |||||||||
| Biological Process | GO:0061470 | T follicular helper cell differentiation | IGI | J:196136 | |||||||||
| Biological Process | GO:0006511 | ubiquitin-dependent protein catabolic process | IBA | J:265628 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/23/2024 MGI 6.23 |

|
|
|
||